Difference between revisions of "Documentation/Nightly/Registration/RegistrationLibrary"
| (41 intermediate revisions by 2 users not shown) | |||
| Line 6: | Line 6: | ||
---- | ---- | ||
| − | *[[Image:UnderConstruction.png| | + | *[[Image:UnderConstruction.png|40px]]Please note this page is under construction and some cases/descriptions may be incomplete. |
| − | *case description: click on the thumbnail image, | + | *case description, step by step instructions, screencasts: click on the thumbnail image, |
*download image data: click on download link, which fetches .mrb file from MIDAS | *download image data: click on download link, which fetches .mrb file from MIDAS | ||
*main module documentation: click on the link in the toolbox column. | *main module documentation: click on the link in the toolbox column. | ||
| Line 19: | Line 19: | ||
! ID | ! ID | ||
! scope="col" width="400px" |Description (click on image to access case page) | ! scope="col" width="400px" |Description (click on image to access case page) | ||
| − | ! Structure | + | ! Structure/Pathology |
! Type of Registration | ! Type of Registration | ||
! Modality | ! Modality | ||
| Line 30: | Line 30: | ||
|Align baseline MR of meningioma with follow-up. | |Align baseline MR of meningioma with follow-up. | ||
|} | |} | ||
| − | | brain tumor | + | | brain / tumor |
| rigid+ affine | | rigid+ affine | ||
| MRI | | MRI | ||
| Line 45: | Line 45: | ||
| MRI | | MRI | ||
|[http://slicer.kitware.com/midas3/download/?items=115374 '''RegLib_C02.mrb'''] <br><small>(input data only , Slicer mrb file. 19 MB) </small><br> | |[http://slicer.kitware.com/midas3/download/?items=115374 '''RegLib_C02.mrb'''] <br><small>(input data only , Slicer mrb file. 19 MB) </small><br> | ||
| − | [http://slicer.kitware.com/midas3/download/ | + | [http://slicer.kitware.com/midas3/download/item/244969/RegLib_C02_full.mrb '''RegLib_C02_full.mrb''']<br><small>(input data + results, Slicer mrb file. 19 MB). </small> |
|- | |- | ||
| align="center" | 3 | | align="center" | 3 | ||
| Line 53: | Line 53: | ||
|Align DTI volume with structural reference scan (T2) | |Align DTI volume with structural reference scan (T2) | ||
|} | |} | ||
| − | | brain tumor | + | | brain / tumor |
| affine + nonrigid | | affine + nonrigid | ||
| DTI | | DTI | ||
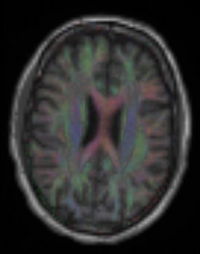
| Line 65: | Line 65: | ||
|Align both intra- and inter-exam MRI of a single subject. | |Align both intra- and inter-exam MRI of a single subject. | ||
|} | |} | ||
| − | | multiple sclerosis | + | | brain / multiple sclerosis |
| affine | | affine | ||
| MRI | | MRI | ||
|[http://slicer.kitware.com/midas3/download/?items=116480 '''RegLib_C04.mrb'''] <br><small>(input data only , Slicer mrb file. 19 MB) </small><br> | |[http://slicer.kitware.com/midas3/download/?items=116480 '''RegLib_C04.mrb'''] <br><small>(input data only , Slicer mrb file. 19 MB) </small><br> | ||
| − | [http://slicer.kitware.com/midas3/download/ | + | [http://slicer.kitware.com/midas3/download/item/244970/RegLib_C04_full.mrb '''RegLib_C04_full.mrb''']<br><small>(input data + results, Slicer mrb file. 19 MB). </small> |
|- | |- | ||
| align="center" | 6 | | align="center" | 6 | ||
| Line 94: | Line 94: | ||
|[http://slicer.kitware.com/midas3/download/?items=116856 '''RegLib_C07.mrb'''] <br><small>(input data only , Slicer mrb file. 8 MB) </small><br> | |[http://slicer.kitware.com/midas3/download/?items=116856 '''RegLib_C07.mrb'''] <br><small>(input data only , Slicer mrb file. 8 MB) </small><br> | ||
[http://slicer.kitware.com/midas3/download/?items=116855 '''RegLib_C07_full.mrb''']<br><small>(input data + results, Slicer mrb file. 27 MB). </small> | [http://slicer.kitware.com/midas3/download/?items=116855 '''RegLib_C07_full.mrb''']<br><small>(input data + results, Slicer mrb file. 27 MB). </small> | ||
| + | |- | ||
| + | | align="center" | 8 | ||
| + | | | ||
| + | {| | ||
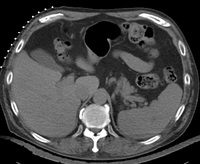
| + | | [[Image:RegLib_C07_Thumb1.png|200px|lleft|RegLib C08: abdominal PET-CT pre/post|link=Documentation:{{documentation/version}}:Registration:RegistrationLibrary:RegLib_C08]] | ||
| + | |Align baseline and follow-up PET/CT pairs | ||
| + | |} | ||
| + | | | ||
| + | | affine+nonrigid | ||
| + | | PET CT | ||
| + | |[http://slicer.kitware.com/midas3/download/?items=116856 '''RegLib_C08.mrb'''] <br><small>(input data only , Slicer mrb file. 8 MB) </small><br> | ||
| + | [http://slicer.kitware.com/midas3/download/?items=116855 '''RegLib_C08_full.mrb''']<br><small>(input data + results, Slicer mrb file. 27 MB). </small> | ||
|- | |- | ||
| align="center" | 9 | | align="center" | 9 | ||
| Line 101: | Line 113: | ||
|Align fMRI with structural MRI | |Align fMRI with structural MRI | ||
|} | |} | ||
| − | | | + | | brain |
| rigid+nonrigid | | rigid+nonrigid | ||
| MRI | | MRI | ||
| Line 113: | Line 125: | ||
|Align probabilistic/group atlas with T1 | |Align probabilistic/group atlas with T1 | ||
|} | |} | ||
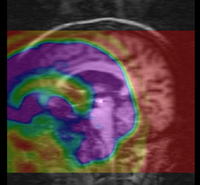
| − | | | + | | brain |
| rigid+nonrigid | | rigid+nonrigid | ||
| MRI | | MRI | ||
| Line 137: | Line 149: | ||
|Align brain PET with low-res MRI | |Align brain PET with low-res MRI | ||
|} | |} | ||
| − | | | + | | brain |
| affine + nonrigid | | affine + nonrigid | ||
| CT | | CT | ||
| Line 149: | Line 161: | ||
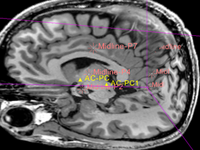
| AC-PC alignment of brain MRI | | AC-PC alignment of brain MRI | ||
|} | |} | ||
| − | | | + | | brain |
| affine + nonrigid | | affine + nonrigid | ||
| CT | | CT | ||
| Line 215: | Line 227: | ||
|- | |- | ||
| align="center" | 32 | | align="center" | 32 | ||
| − | | | + | | |
{| | {| | ||
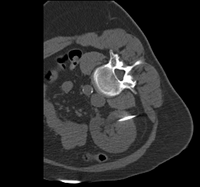
| [[Image:RegLib_C32_Thumb4.png|200px|left|link=Documentation:{{documentation/version}}:Registration:RegistrationLibrary:RegLib_C32]] | | [[Image:RegLib_C32_Thumb4.png|200px|left|link=Documentation:{{documentation/version}}:Registration:RegistrationLibrary:RegLib_C32]] | ||
| Alignment of structural reference T1 to a DTI scan | | Alignment of structural reference T1 to a DTI scan | ||
|} | |} | ||
| − | | | + | | brain |
| affine + nonrigid | | affine + nonrigid | ||
| DTI | | DTI | ||
|[http://slicer.kitware.com/midas3/download/?items=103594 '''RegLib_C32.mrb'''] <small>(input data, Slicer mrb file. 31 MB). </small> <br> | |[http://slicer.kitware.com/midas3/download/?items=103594 '''RegLib_C32.mrb'''] <small>(input data, Slicer mrb file. 31 MB). </small> <br> | ||
[http://slicer.kitware.com/midas3/download/?items=103593 '''RegLib_C32_full.mrb'''] <small>(input data+results, Slicer mrb file. 36 MB). </small> | [http://slicer.kitware.com/midas3/download/?items=103593 '''RegLib_C32_full.mrb'''] <small>(input data+results, Slicer mrb file. 36 MB). </small> | ||
| + | |- | ||
| + | | align="center" | 37 [[Image:UnderConstruction.png|40px]] | ||
| + | | | ||
| + | {| | ||
| + | | [[Image:RegLib_C37_Thumb1.png|200px|lleft|RegLib C37: tumor resection|link=Documentation:{{documentation/version}}:Registration:RegistrationLibrary:RegLib_C37]] | ||
| + | | brain tumor resection | ||
| + | |} | ||
| + | | brain | ||
| + | | non-rigid | ||
| + | | MRI | ||
| + | |[http://slicer.kitware.com/midas3/download/?items=xx '''RegLib_C37.mrb'''] <br><small>(input data only, Slicer mrb file. 12 MB) </small><br> | ||
| + | [http://slicer.kitware.com/midas3/download/?items=xx '''RegLib_C7_full.mrb''']<br><small>(input data + results, Slicer mrb file. 36 MB). </small> | ||
|- | |- | ||
| align="center" | 38 | | align="center" | 38 | ||
| Line 236: | Line 260: | ||
|MRI | |MRI | ||
|[http://slicer.kitware.com/midas3/download/?items=103791 '''RegLib_C38.mrb'''] <small>(input data, Slicer mrb file. 42 MB). </small> <br> | |[http://slicer.kitware.com/midas3/download/?items=103791 '''RegLib_C38.mrb'''] <small>(input data, Slicer mrb file. 42 MB). </small> <br> | ||
| + | |- | ||
| + | | align="center" | 39 | ||
| + | | | ||
| + | {| | ||
| + | | [[Image:RegLib_C39_Thumb1.png|200px|lleft|RegLib C39: in vitro CT of human vertebra|link=Documentation:{{documentation/version}}:Registration:RegistrationLibrary:RegLib_C39]] | ||
| + | | in vitro CT of human vertebra before and after structural damage | ||
| + | |} | ||
| + | | vertebra | ||
| + | | rigid | ||
| + | | CT | ||
| + | |[http://slicer.kitware.com/midas3/download/bitstream/431015/RegLib_C39.mrb '''RegLib_C39.mrb'''] <br><small>(input data only, Slicer mrb file. 25 MB) </small><br> | ||
| + | [http://slicer.kitware.com/midas3/download/bitstream/153889/RegLib_C39_full.mrb '''RegLib_C39_full.mrb''']<br><small>(input data + results, Slicer mrb file. 45 MB). </small> | ||
| + | |- | ||
| + | | align="center" | 40 | ||
| + | | | ||
| + | {| | ||
| + | | [[Image:RegLib_C40_Thumb1.png|200px|lleft|RegLib C40: micro CT of material sample|link=Documentation:{{documentation/version}}:Registration:RegistrationLibrary:RegLib_C40]] | ||
| + | | register micro-CT of wood sample to quantify/model structural changes | ||
| + | |} | ||
| + | | wood sample | ||
| + | | non-rigid | ||
| + | | micro-CT | ||
| + | |[http://slicer.kitware.com/midas3/download/?items=119692 '''RegLib_C40.mrb'''] <br><small>(input data only, Slicer mrb file. 32 MB) </small><br> | ||
| + | [http://slicer.kitware.com/midas3/download/?items=119691 '''RegLib_C40_full.mrb''']<br><small>(input data + results, Slicer mrb file. 100 MB). </small> | ||
| + | |- | ||
| + | | align="center" | 41 | ||
| + | | | ||
| + | {| | ||
| + | | [[Image:RegLib_C41_Thumb1.png|200px|lleft|RegLib C41: mouse brain MRI|link=Documentation:{{documentation/version}}:Registration:RegistrationLibrary:RegLib_C41]] | ||
| + | | Align two distinct mouse brain MRIs | ||
| + | |} | ||
| + | | mouse brain | ||
| + | | non-rigid | ||
| + | | MRI | ||
| + | |[http://slicer.kitware.com/midas3/download/?items=119689 '''RegLib_C41.mrb'''] <br><small>(input data only, Slicer mrb file. 32 MB) </small><br> | ||
| + | [http://slicer.kitware.com/midas3/download/?items=119688 '''RegLib_C41_full.mrb''']<br><small>(input data + results, Slicer mrb file. 100 MB). </small> | ||
| + | |- | ||
| + | | align="center" | 42 | ||
| + | | | ||
| + | {| | ||
| + | | [[Image:RegLib_C42_Thumb1.png|200px|lleft|RegLib C42: pediatric MRI|link=Documentation:{{documentation/version}}:Registration:RegistrationLibrary:RegLib_C42]] | ||
| + | | autism | ||
| + | |} | ||
| + | | brain | ||
| + | | non-rigid | ||
| + | | MRI | ||
| + | |[http://slicer.kitware.com/midas3/download/?items=119557 '''RegLib_C42.mrb'''] <br><small>(input data only, Slicer mrb file. 12 MB) </small><br> | ||
| + | [http://slicer.kitware.com/midas3/download/?items=119556 '''RegLib_C42_full.mrb''']<br><small>(input data + results, Slicer mrb file. 36 MB). </small> | ||
| + | |- | ||
| + | | align="center" | 44 | ||
| + | | | ||
| + | {| | ||
| + | | [[Image:RegLib_C44_Thumb1.png|200px|lleft|RegLib C44: pelvic MRI|link=Documentation:{{documentation/version}}:Registration:RegistrationLibrary:RegLib_C44]] | ||
| + | | pelvis | ||
| + | |} | ||
| + | | abdominal | ||
| + | | non-rigid | ||
| + | | MRI | ||
| + | |[http://slicer.kitware.com/midas3/download/?items=119555 '''RegLib_C44.mrb'''] <br><small>(input data only, Slicer mrb file. 74 MB) </small><br> | ||
| + | [http://slicer.kitware.com/midas3/download/?items=119554 '''RegLib_C44_full.mrb''']<br><small>(input data + results, Slicer mrb file. 82 MB). </small> | ||
| + | |- | ||
| + | | align="center" | 46 | ||
| + | | | ||
| + | {| | ||
| + | | [[Image:RegLib_C46_DisplacementField.gif|200px|lleft|RegLib C46: 2D cine MRI of breathing cycle|link=Documentation:{{documentation/version}}:Registration:RegistrationLibrary:RegLib_C46]] | ||
| + | | 2D cine MRI of breathing cycle | ||
| + | |} | ||
| + | | abdominal | ||
| + | | non-rigid 2D , batch processing | ||
| + | | MRI | ||
| + | |[http://slicer.kitware.com/midas3/download/?items=119459 '''RegLib_C46.mrb'''] <br><small>(input data (ref + 1 slice only), Slicer mrb file. 0.6 MB) </small><br> | ||
| + | [http://slicer.kitware.com/midas3/download/?items=119460 '''RegLib_C46_Data.zip''']<br><small>(input data + results, zip file with original DICOM + registration script. 65 MB). </small> | ||
| + | |- | ||
| + | | align="center" | 48 | ||
| + | | | ||
| + | {| | ||
| + | | [[Image:RegLib_C48_Thumb3.png|200px|lleft|RegLib C48: Fiducial Registration of Abdominal MR-CT|link=Documentation:{{documentation/version}}:Registration:RegistrationLibrary:RegLib_C48]] | ||
| + | | Preop MRI to intraop interventional CT | ||
| + | |} | ||
| + | | abdominal | ||
| + | | Fiducial-based rigid | ||
| + | | CT | ||
| + | |[http://slicer.kitware.com/midas3/download/?items=118088 '''RegLib_C48.mrb'''] <br><small>(input data only (incl. example fiducials) , Slicer mrb file. 10 MB) </small><br> | ||
| + | [http://slicer.kitware.com/midas3/download/?items=118087 '''RegLib_C48_full.mrb''']<br><small>(input data + results, Slicer mrb file. 12 MB). </small> | ||
| + | |- | ||
| + | | align="center" | 49 | ||
| + | | | ||
| + | {| | ||
| + | | [[Image:RegLib_C49_Thumb1.png|200px|lleft|RegLib C49: Pediatric Brain MR-DTI|link=Documentation:{{documentation/version}}:Registration:RegistrationLibrary:RegLib_C49]] | ||
| + | | Pediatric Brain MR-DTI | ||
| + | |} | ||
| + | | brain | ||
| + | | affine + BSpline | ||
| + | | MRI | ||
| + | |[http://slicer.kitware.com/midas3/download/?items=118347 '''RegLib_C49.mrb'''] <br><small>(input data only (incl. example fiducials) , Slicer mrb file. 50 MB) </small><br> | ||
| + | [http://slicer.kitware.com/midas3/download/?items=118346 '''RegLib_C49_full.mrb''']<br><small>(input data + results, Slicer mrb file. 82 MB). </small> | ||
| + | |- | ||
| + | | align="center" | 50 | ||
| + | | | ||
| + | {| | ||
| + | | [[Image:RegLib_C50_Thumb1.png|200px|lleft|RegLib C50: Mouse CT|link=Documentation:{{documentation/version}}:Registration:RegistrationLibrary:RegLib_C50]] | ||
| + | | mouse brain micro-CT | ||
| + | |} | ||
| + | | mouse brain | ||
| + | | affine + BSpline | ||
| + | | micro-CT | ||
| + | |[http://slicer.kitware.com/midas3/download/?items=127956 '''RegLib_C50.mrb'''] <br><small>(input data only (incl. example fiducials) , Slicer mrb file. 50 MB) </small><br> | ||
| + | [http://slicer.kitware.com/midas3/download/?items=127955 '''RegLib_C50_full.mrb''']<br><small>(input data + results, Slicer mrb file. 82 MB). </small> | ||
|} | |} | ||
<!-- | <!-- | ||
this is commented out | this is commented out | ||
--> | --> | ||
Latest revision as of 20:29, 14 May 2018
Home < Documentation < Nightly < Registration < RegistrationLibrary
|
For the latest Slicer documentation, visit the read-the-docs. |
- Slicer Registration FAQ: answers to the most common questions/problems, including cookbook recipes for common registration steps.
- Slicer Registration Training Videos: brief videos/screencasts showing step-by-step approaches to the most common tasks
- Slicer Resampling Overview a summary of how 3DSlicer handles transforms and how image resampling is performed.]]
- case description, step by step instructions, screencasts: click on the thumbnail image,
- download image data: click on download link, which fetches .mrb file from MIDAS
- main module documentation: click on the link in the toolbox column.
- sort the table by a category: click on the sort icon in the column header
| ID | Description (click on image to access case page) | Structure/Pathology | Type of Registration | Modality | Download | ||
|---|---|---|---|---|---|---|---|
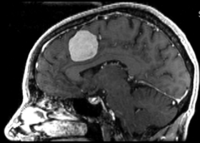
| 1 |
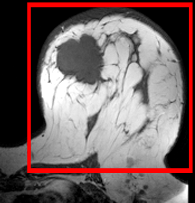
|
brain / tumor | rigid+ affine | MRI | RegLib_C01.mrb (input data only , Slicer mrb file. 13 MB) | ||
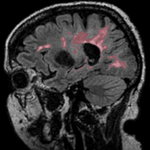
| 2 |
|
multiple sclerosis | affine | MRI | RegLib_C02.mrb (input data only , Slicer mrb file. 19 MB) RegLib_C02_full.mrb | ||
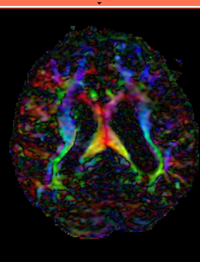
| 3 |
|
brain / tumor | affine + nonrigid | DTI | RegLib_C03.mrb (input data only , Slicer mrb file. 50 MB) RegLib_C03_full.mrb | ||
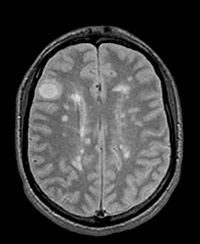
| 4 |
|
brain / multiple sclerosis | affine | MRI | RegLib_C04.mrb (input data only , Slicer mrb file. 19 MB) RegLib_C04_full.mrb | ||
| 6 |
|
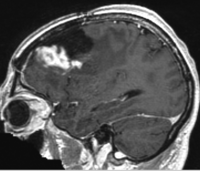
breast cancer | affine+nonrigid | MRI | RegLib_C06.mrb (input data only , Slicer mrb file. 40 MB) RegLib_C06_full.mrb | ||
| 7 |
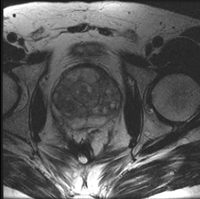
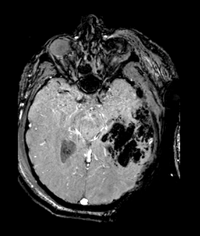
|
prostate cancer | affine+nonrigid | MRI | RegLib_C07.mrb (input data only , Slicer mrb file. 8 MB) RegLib_C07_full.mrb | ||
| 8 |
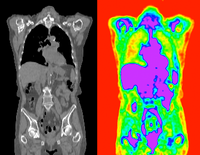
|
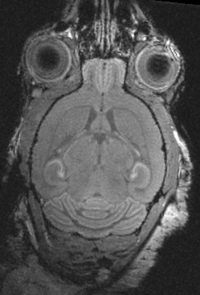
affine+nonrigid | PET CT | RegLib_C08.mrb (input data only , Slicer mrb file. 8 MB) RegLib_C08_full.mrb | |||
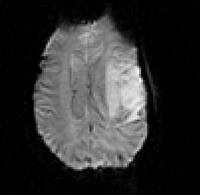
| 9 |
|
brain | rigid+nonrigid | MRI | RegLib_C09.mrb (input data only , Slicer mrb file. 40 MB) RegLib_C09_full.mrb | ||
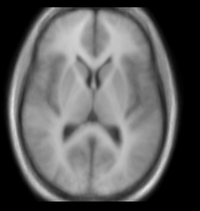
| 10 |
|
brain | rigid+nonrigid | MRI | RegLib_C10.mrb (input data only , Slicer mrb file. 12 MB) RegLib_C10_full.mrb | ||
| 12 |
|
liver tumor | affine + nonrigid | CT | RegLib_C12.mrb (input data only , Slicer mrb file. 35 MB) RegLib_C12_full.mrb | ||
| 14 |
|
brain | affine + nonrigid | CT | RegLib_C14.mrb (input data only , Slicer mrb file. 9 MB) RegLib_C14_full.mrb | ||
| 15 |
|
brain | affine + nonrigid | CT | RegLib_C15.mrb (input data only (incl. example fiducials) , Slicer mrb file. 22 MB) RegLib_C15_full.mrb | ||
| 17 |
|
abdominal | affine | CT | RegLib_C17.mrb (input data only (incl. example fiducials) , Slicer mrb file. 10 MB) RegLib_C17_full.mrb | ||
| 20 |
|
full body | rigid + affine + nonrigid | PET/CT | RegLib_C20.mrb (input data only , Slicer mrb file. 76 MB) RegLib_C20_full.mrb | ||
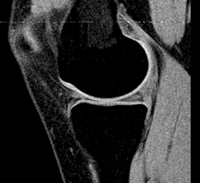
| 21 |
|
knee | rigid + affine + nonrigid | MRI | RegLib_C21.mrb (input data + results, Slicer mrb file. 12 MB). | ||
| 27 |
|
brain tumor | affine + nonrigid | DTI | RegLib_C27.mrb (input data, Slicer mrb file 50MB) RegLib_C27_full.mrb | ||
| 29 |
|
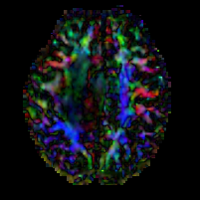
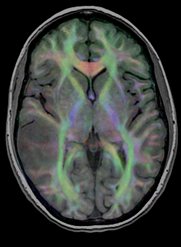
brain tumor | affine + nonrigid | DTI | RegLib_C29_raw.mrb (input data, Slicer mrb file. 71 MB). RegLib_C29_full.mrb | ||
| 32 |
|
brain | affine + nonrigid | DTI | RegLib_C32.mrb (input data, Slicer mrb file. 31 MB). RegLib_C32_full.mrb (input data+results, Slicer mrb file. 36 MB). | ||
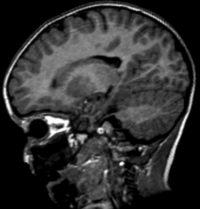
| 37 |
|
brain | non-rigid | MRI | RegLib_C37.mrb (input data only, Slicer mrb file. 12 MB) RegLib_C7_full.mrb | ||
| 38 |
|
traumatic brain injury (TBI) | rigid | MRI | RegLib_C38.mrb (input data, Slicer mrb file. 42 MB). | ||
| 39 |
|
vertebra | rigid | CT | RegLib_C39.mrb (input data only, Slicer mrb file. 25 MB) RegLib_C39_full.mrb | ||
| 40 |
|
wood sample | non-rigid | micro-CT | RegLib_C40.mrb (input data only, Slicer mrb file. 32 MB) RegLib_C40_full.mrb | ||
| 41 |
|
mouse brain | non-rigid | MRI | RegLib_C41.mrb (input data only, Slicer mrb file. 32 MB) RegLib_C41_full.mrb | ||
| 42 |
|
brain | non-rigid | MRI | RegLib_C42.mrb (input data only, Slicer mrb file. 12 MB) RegLib_C42_full.mrb | ||
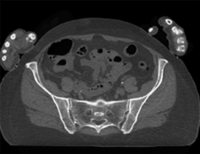
| 44 |
|
abdominal | non-rigid | MRI | RegLib_C44.mrb (input data only, Slicer mrb file. 74 MB) RegLib_C44_full.mrb | ||
| 46 |
|
abdominal | non-rigid 2D , batch processing | MRI | RegLib_C46.mrb (input data (ref + 1 slice only), Slicer mrb file. 0.6 MB) RegLib_C46_Data.zip | ||
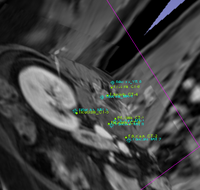
| 48 |
|
abdominal | Fiducial-based rigid | CT | RegLib_C48.mrb (input data only (incl. example fiducials) , Slicer mrb file. 10 MB) RegLib_C48_full.mrb | ||
| 49 |
|
brain | affine + BSpline | MRI | RegLib_C49.mrb (input data only (incl. example fiducials) , Slicer mrb file. 50 MB) RegLib_C49_full.mrb | ||
| 50 |
|
mouse brain | affine + BSpline | micro-CT | RegLib_C50.mrb (input data only (incl. example fiducials) , Slicer mrb file. 50 MB) RegLib_C50_full.mrb |