Modules:FourDAnalysis-Documentation-3.6
Return to Slicer 3.6 Documentation
Currently, there is no binary distribution for 4D Analysis module available, since we are have issues in building SciPy, which the module depends on. If you would like to try the module, please refer Development section bellow for further information.
Module Name
4D Analysis Module
General Information
Module Type & Category
Type: Interactive
Category: 4D
Authors, Collaborators & Contact
- Junichi Tokuda, Ph.D., Brigham and Women's Hospital (Developer)
- Wendy J Plesniak, Ph.D., Brigham and Women's Hospital (Developer)
- Hatsuho Mamata, M.D., Ph.D., Brigham and Women's Hospital (Clinical)
- Hiroto Hatabu, M.D., Ph.D., Brigham and Women's Hospital (Clinical)
- Ron Kikinis, M.D., Brigham and Women's Hospital (Advisor)
- Robert Lenkinski, Ph.D., Beth Israel Deaconess Medical Center (Advisor)
Contact: Junichi Tokuda, tokuda at bwh.harvard.edu
Module Description
The 4D Analysis Module allows you to analyze a time-series of 3D images (4D image) on 3D Slicer. Currently, the module offers the following features:
- Intensity plot. The module can draw intensity curves at the specified regions of interest.
- Pharmacokinetic model analysis. (SciPy required) The users can perform pharmacokinetic model analysis based on the intensity curves. Models are defined in Python scripts so that the users can modify the existing models or create their own pharmacokinetic models.
- Parameter map. (SciPy required) The module also allows generating a map of pharmacokinetic parameters.
In default, only the intensity plot is available. To use the other functions, you need to install scipy for 3D Slicer and recompile 4D Analysis module. Please refer the section bellow for more detail. Slicer3+SciPy installation memo has some tips regarding SciPy installation in 3D Slicer for Mac OS X.
Usage
Quick Tour of Features and Use
Basic features
- Active 4D Bundle: Specify the active 4D image.
- Frame Control: View the 4D image data. 4D images can be loaded from 4D Image Module.
- Intensity Plot: Generate intensity curves and display them on this panel. The curves are measured for each labeled region in the specified label map. If the label map contains multiple labels, the module generates intensity curve for each label. The user can also load/save curves from/to CSV files. The plotting graph can be controlled from Python interactor or other 3D Slicer modules. See Slicer3:2DPlotting for more detail.
Advanced features (require SciPy installation)
- Model/Parameter: Select Python script and parameters for pharmacokinetic analysis.
- Curve Fitting: Perform a single curve fitting. It displays the result parameters.
- Parameter Map: Generate parameter map. The module performs curve fitting for each voxel in the specified region.
Tutorial 1: Plotting time-intensity curve from 4D image data
The module allows plotting mean intensities in the specified region on the series of images.
Step 1: Load 4D image
Please follow 4D Image module documentation page to load time-series image data.
Step 2: Define a mask
After loading 4D image from 4D Image module, open Editor and make a label map to mask regions you want to measure mean intensities. When you make a label map, please specify one of display volume nodes created by 4D Image module (a volume node with a name "*_Display1") (Figure 2-3).
Step 3: Plot
Open 4D Analysis module in 4D category, and choose the 4D image loaded in the first step from Active 4D Bundle node selector menu (Figure 4). If you have only one 4D image loaded on your Slicer, it should be already there.
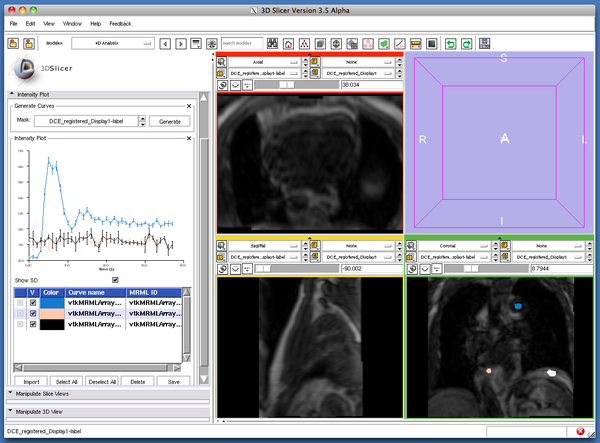
After specifying the 4D image, open Intensity Plot tab and select the mask volume, which you created in the previous step (Figure 5), and then press Generate' button. If you have multiple labels in the label volume, the module generates time-intensity curves for each label (Figure 6).
Step 4: Show and hide curves
The column list bellow the graph allows you to show and hide the curves by clicking the check boxes in the V column.
Step 5: Save intensity curve data
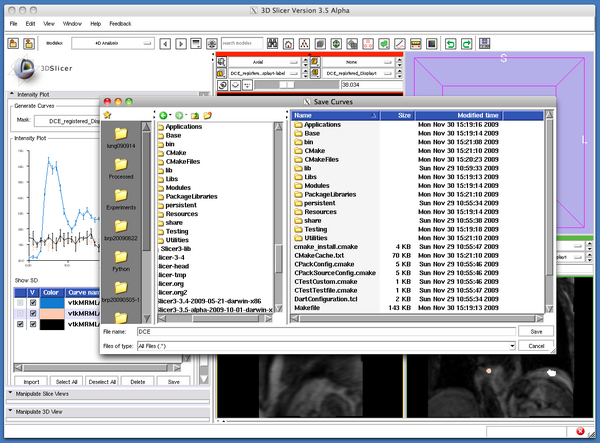
To save the plot data as CSV files, turn on the check boxes in the left column of the curves to save in the column list, and then click the Save button bellow the column list (Figure 8). The Slicer will pop up a dialog window (Figure 9). The curve data is stored in CSV files, which can be loaded to 4D Analysis Module later.
Step 6: Delete intensity curve data
To delete the plot data, turn on the check boxes in the left column of the curves to delete in the column list, and then click the Delete button bellow the column list (Figure 10). The Slicer will pop up a confirmation dialog box(Figure 11).
Development
Installing 4D Analysis Module
The following instruction describes how to install the 4D Analysis with basic functions (curve plotting). This is also necessary to set curve analysis described in the next section.
Step 1: Put source directory in the modules directory
$ cd <working dir>/Slicer3/Modules/ $ svn co http://svn.na-mic.org/NAMICSandBox/trunk/FourDAnalysis
Step 2: Edit the CMakeLists.txt
Edit the CMakeLists.txt in <working dir>/Slicer3/Modules/ and add FourDAnalysis around line 86 in SET(modules ...) section.
Step 3: Go to the build directory and run make
$ cd <working dir>/Slicer3-build/Modules/ $ make
This will updates all CMake cache appropriately and build the 4D Analysis module. After the process, please start 3D Slicer and check if 4D Analysis module is in the module menu (it should be in the "4D" section).
Setting up curve analysis
The module allows you to fit a model to actual time-intensity curves obtained from DCE MRI. Since this function requires SciPy package, which does not come with 3D Slicer in default, you need to install SciPy in 3D Slicer.
Step 1: Install SciPy on Slicer 3
Please refer Slicer3 Python page. See Slicer3+SciPy installation memo for Mac OS X user (the memo was updated on Apr, 2011).
Step 2: Building 4D Analysis module with SciPy Option
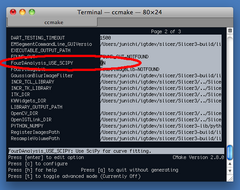
Once you set up 3D Slicer with SciPy, you can set FourDAnalysis_USE_SCIPY "ON" by running make with edit_cache option:
$ cd <Slicer3-build directory> $ cd Modules/FourDAnalysis $ make edit_cache
Make program will launch ccmake (full screen CMake configuration tool) (Figure 12), where you can turn on FourDAnalysis_USE_SCIPY macro. After generating a new make file, quit ccmake screen and run make.
After building the module, run 3D Slicer to check if the 4D Analysis module has been built with SciPy option.
Dependencies
The intensity curve analysis functions depend on SciPy package. Please turn on SciPy option when you build your 3D Slicer.
Known bugs
Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Customize following links for your module.
Links to documentation generated by doxygen.
More Information
2D Graph
The module provides a set of classes to support 2D Graph for intensity curve plotting, which can potentially be used for other projects. Please refer Slicer3:2DPlotting for more detail.
Acknowledgment
This work is supported by NIH (5R21CA116271: Dynamic contrast-enhanced MRI of pulmonary nodule at 3T, PI: Hiroto Hatabu), NA-MIC, and NCIGT.
References
Sample DCE data set and after conversion to NRRD. This is from a prostate DCE acquisition.