Documentation/4.6/Modules/FiberBundleToLabelMap
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Author(s)/Contributor(s): Steve Pieper (SPL, Isomics, Inc.), Isaiah Norton (SPL, LMI, BWH, SlicerDMRI) License: 3D Slicer Contribution and Software License Agreement Acknowledgements: The SlicerDMRI developers gratefully acknowledge funding for this project provided by NIH NCI ITCR U01CA199459 (Open Source Diffusion MRI Technology For Brain Cancer Research), NIH P41EB015898 (National Center for Image-Guided Therapy) and NIH P41EB015902 (Neuroimaging Analysis Center), as well as the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Contact: slicer-users@bwh.harvard.edu Website: http://slicerdmri.github.io/ | |||||||||||
|
Module Description
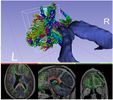
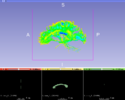
This module sets the specified label value in the label map at every vertex in each of the fibers in a bundle.
This module first upsamples points along the fiber bundle in order to get better voxel coverage.
Tutorials
Panels and their use
- Fiber Bundle
- Pick a fiber bundle to rasterize
- Target Label Map
- The fibers will be painted into this label map. Note that this will add to and overwrite existing data, but does not clear the label map to zero first.
- Label Value
- Numerical value to be written into the label map.
Similar Modules
References
Information for Developers
This is a python scripted module.