Documentation/4.5/Modules/SobolevSegmenter
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research through grant U54 EB005149. Information on NA-MIC can be obtained from the NA-MIC website. | |||
|
Module Description
This extension implements Sobolev inner product based active contour, using the Chan-Vese energy functional. The segmentation is appropriate for 2D images. Extensions are being made now to the 3D case and will be available in the near future. The parametric contour is generally smooth, and able to capture concavities.
Use Cases
The Sobolev segmenter is a general image segmenter, and it can be used with any type of data, as explained in the tutorial.
Tutorials
The 2D image segmentation:
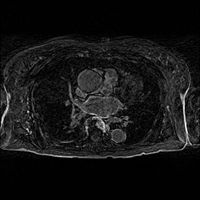
- Load the image (input volume):
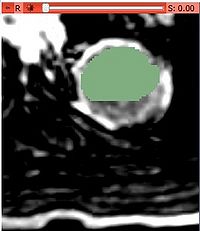
- Use built in editor to select a single initial mask (or load a binary mask file):
- Select Segmenation->SobolevSegmenter module
- Choose the Input Volume and the Initial Mask accordingly. Create a new volume for the Output Volume.
- Press Apply button.
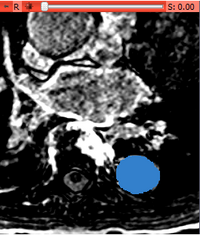
- After a few second the following output volume should appear:
The 3D volume segmentation tutorial: [1]
Panels and their use
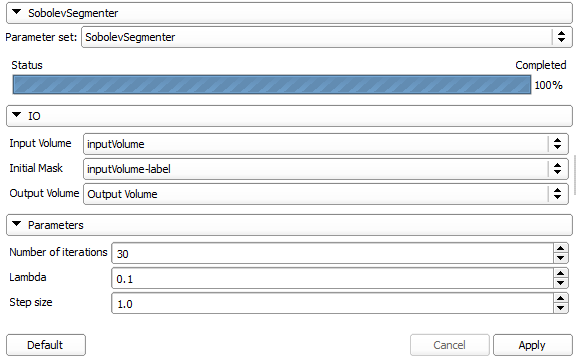
The module has the following panel:
The IO section of this panel defines two input images (data and initial mask) and one output image (final mask). The algorithm has three parameters: self-explanatory number of iterations and contour evolution step size. In addition, the parameter lambda chooses the smoothness of the contour (smoothing kernel width).
Similar Modules
N/A
References
N/A
Information for Developers
| Section under construction. |