Difference between revisions of "Documentation:Nightly:Registration:RegistrationLibrary:RegLib C42"
| Line 12: | Line 12: | ||
|} | |} | ||
| − | == Modules | + | == Modules used == |
| + | *[[Documentation/Nightly/Modules/BRAINSFit| ''General Registration (BRAINS)'']] | ||
| + | |||
| + | == Download (from NAMIC MIDAS) == | ||
| + | <small>''Why 2 sets of files? The "input data" mrb includes only the unregistered data to try the method yourself from start to finish. The full dataset includes intermediate files and results (transforms, resampled images etc.). If you use the full dataset we recommend to choose different names for the images/results you create yourself to distinguish the old data from the new one you generated yourself. ''</small> | ||
| + | *[http://slicer.kitware.com/midas3/download/?items=119557 '''RegLib_C42.mrb'''] <br><small>(input data only, Slicer mrb file. 12 MB) </small><br> | ||
| + | *[http://slicer.kitware.com/midas3/download/?items=119556 '''RegLib_C42_full.mrb''']<br><small>(input data + results, Slicer mrb file. 36 MB). </small> | ||
== Description == | == Description == | ||
This is a typical case of change assessment. | This is a typical case of change assessment. | ||
| − | |||
| − | |||
| − | |||
=== Keywords === | === Keywords === | ||
MRI, brain, head, intra-subject, T1, autism, change assessment, pediatric MRI | MRI, brain, head, intra-subject, T1, autism, change assessment, pediatric MRI | ||
| − | + | == Procedure == | |
| − | == | ||
*'''Phase I: Register Rigid/Affine''' | *'''Phase I: Register Rigid/Affine''' | ||
#open Registration : ''BrainsFit'' module (presets: BRAINSfit_Xf1 or _Xf2 | #open Registration : ''BrainsFit'' module (presets: BRAINSfit_Xf1 or _Xf2 | ||
| Line 41: | Line 43: | ||
##Leave all other settings at default | ##Leave all other settings at default | ||
##click Apply | ##click Apply | ||
| − | |||
| + | == Registration Results== | ||
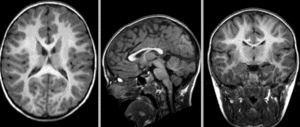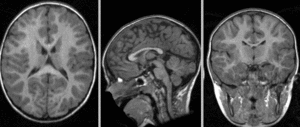
[[Image:RegLib_C42_unregistered.gif|300px]] baseline and follow-up before registration (click to enlarge) <br> | [[Image:RegLib_C42_unregistered.gif|300px]] baseline and follow-up before registration (click to enlarge) <br> | ||
[[Image:RegLib_C42_affine.gif|300px]] after affine alignment. Note the residual shape differences (click to enlarge) <br> | [[Image:RegLib_C42_affine.gif|300px]] after affine alignment. Note the residual shape differences (click to enlarge) <br> | ||
Revision as of 18:57, 1 October 2013
Home < Documentation:Nightly:Registration:RegistrationLibrary:RegLib C42Contents
Slicer Registration Library Case #42:
Intra-subject Brain MRI: serial assessment of brain development in autism
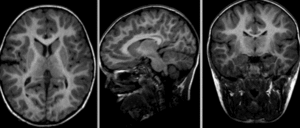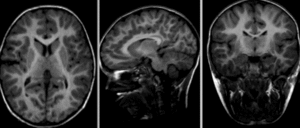
Input
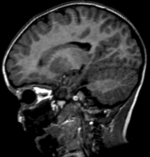
|

|
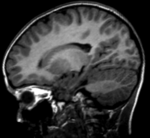
|
| fixed image/target | moving image |
Modules used
Download (from NAMIC MIDAS)
Why 2 sets of files? The "input data" mrb includes only the unregistered data to try the method yourself from start to finish. The full dataset includes intermediate files and results (transforms, resampled images etc.). If you use the full dataset we recommend to choose different names for the images/results you create yourself to distinguish the old data from the new one you generated yourself.
- RegLib_C42.mrb
(input data only, Slicer mrb file. 12 MB) - RegLib_C42_full.mrb
(input data + results, Slicer mrb file. 36 MB).
Description
This is a typical case of change assessment.
Keywords
MRI, brain, head, intra-subject, T1, autism, change assessment, pediatric MRI
Procedure
- Phase I: Register Rigid/Affine
- open Registration : BrainsFit module (presets: BRAINSfit_Xf1 or _Xf2
- Input Parameters:
- Fixed: "Autism_T1_2yrs"
- Moving: "Autism_T1_4yrs"
- Registration Phases:
- select/check Include Rigid registration phase , Include Scale Versor 3D , Include Affine, Include BSpline
- Output Settings
- select a new "Slicer BSpline Transform", rename to "Xf3_Affine+BSpline" or similar "
- select a new volume "Output Image Volume, rename to "Autism_T1_4yrs_Xf3", indicating that this is the result volume after being resampled with the Xf3 transform.
- Output Image Pixel Type: check "ushort". As a general rule, match the datatype of the input image, unless specific reason demands otherwise (e.g. you wish to avoid rounding of interpolation effects). The "float" datatype is 4-bytes per pixel and can produce unnecessary large files.
- Registration Parameters:
- increase Number Of Samples to 300,000
- Registration Parameters: set Number Of Grid Subdivisions to 5,5,5
- Leave all other settings at default
- click Apply
- Input Parameters:
Registration Results
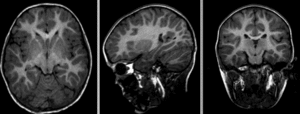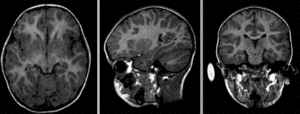 baseline and follow-up before registration (click to enlarge)
baseline and follow-up before registration (click to enlarge)
 after affine alignment. Note the residual shape differences (click to enlarge)
after affine alignment. Note the residual shape differences (click to enlarge)
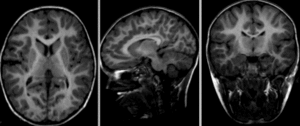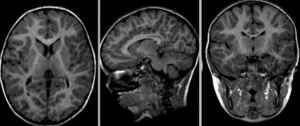 after nonrigid BSpline registration (click to enlarge)
after nonrigid BSpline registration (click to enlarge)
 | visualization of the nonrigid deformation component only
| visualization of the nonrigid deformation component only