Difference between revisions of "Documentation/Nightly/Extensions/LesionSimulator"
Acsenrafilho (talk | contribs) m |
Acsenrafilho (talk | contribs) m Tag: 2017 source edit |
||
| (8 intermediate revisions by the same user not shown) | |||
| Line 10: | Line 10: | ||
This work was partially funded by CAPES and CNPq, a Brazillian Agencies. Information on CAPES can be obtained on the [http://www.capes.gov.br/ CAPES website] and [http://www.cnpq.br/ CNPq website].<br> | This work was partially funded by CAPES and CNPq, a Brazillian Agencies. Information on CAPES can be obtained on the [http://www.capes.gov.br/ CAPES website] and [http://www.cnpq.br/ CNPq website].<br> | ||
Authors: Antonio Carlos da S. Senra Filho and Fabrício Henrique Simozo, CSIM Laboratory (University of Sao Paulo, Department of Computing and Mathematics)<br> | Authors: Antonio Carlos da S. Senra Filho and Fabrício Henrique Simozo, CSIM Laboratory (University of Sao Paulo, Department of Computing and Mathematics)<br> | ||
| − | + | Contacts: Antonio Carlos da S. Senra Filho <email>acsenrafilho@usp.br</email> - Fabrício Henrique Simozo <email>fsimozo@gmail.com</email><br> | |
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
{{documentation/{{documentation/version}}/module-introduction-logo-gallery | {{documentation/{{documentation/version}}/module-introduction-logo-gallery | ||
| Line 27: | Line 27: | ||
[[Image:LesionSimulator-logo.png|left]] | [[Image:LesionSimulator-logo.png|left]] | ||
| − | This extension offer a set of tools for brain lesion simulation, based on MRI images. At moment, the module MS Lesion Simulator is available, where it can simulates both baseline scan lesion volumes (given a lesion load) and longitudinal image simulations. In summary, a statistical lesion database is generated based on a set of manual lesion mark-ups, being non-linearly registered to MNI152 space (isotropic 1mm of voxel resolution). Using a small set of parameters (lesion load, lesion homogeneity, lesion intensity indenpendence and lesion variability), it is possible to generate a broad range of MS lesions patterns in multimodal MRI imaging techniques (at moment, T1, T2, T2-FLAIR, PD, DTI-FA and DTI-ADC images are provided). For more details about this project, please see the [http://dx.doi.org/ original paper]. | + | This extension offer a set of tools for brain lesion simulation, based on MRI images. At moment, the module MS Lesion Simulator is available, where it can simulates both baseline scan lesion volumes (given a lesion load) and longitudinal image simulations. In summary, a statistical lesion database is generated based on a set of manual lesion mark-ups, being non-linearly registered to MNI152 space (isotropic 1mm of voxel resolution), and a local contrast ratio is applied on each image modality provided by the user. Using a small set of parameters (lesion load, lesion homogeneity, lesion intensity indenpendence and lesion variability), it is possible to generate a broad range of MS lesions patterns in multimodal MRI imaging techniques (at moment, T1, T2, T2-FLAIR, PD, DTI-FA and DTI-ADC images are provided). For more details about this project, please see the [http://dx.doi.org/10.1088/2057-1976/ab08fc original paper]. |
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|Modules}} | {{documentation/{{documentation/version}}/extension-section|Modules}} | ||
| − | *[[Documentation/{{documentation/version}}/Modules/|MS | + | *[[Documentation/{{documentation/version}}/Modules/MSLesionSimulator|MS Lesion Simulator]] |
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 41: | Line 39: | ||
**In the baseline scan approach, it is offered a simulation procedure where a determined lesion load is reconstructed using a subject specific anatomical features, resulting in a realistic MS lesion load simulation. | **In the baseline scan approach, it is offered a simulation procedure where a determined lesion load is reconstructed using a subject specific anatomical features, resulting in a realistic MS lesion load simulation. | ||
* Use Case 2: Simulate longitudinal MS lesion progression on clinical healthy individuals MRI images | * Use Case 2: Simulate longitudinal MS lesion progression on clinical healthy individuals MRI images | ||
| − | ** | + | **Another important issue in Multiple Sclerosis diagnosis is the lesion progression, where drive the therapeutic strategy by the health professionals. In this case, an automatic segmentation approach could be optimally adjusted for the time progression presented in a exam series. |
<gallery widths="300px" heights="300px" perrow="2"> | <gallery widths="300px" heights="300px" perrow="2"> | ||
| Line 52: | Line 50: | ||
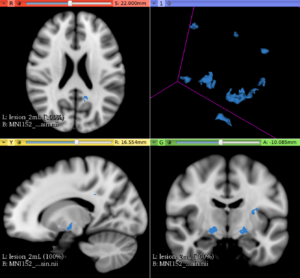
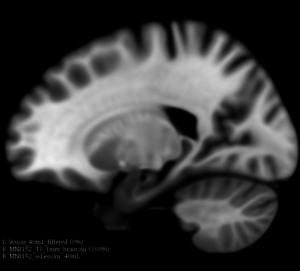
Image:MNI152_orig_sag.png|T1 weighted MRI brain in sagittal orientation (provided by the ICBM-MNI152 non linear brain template) | Image:MNI152_orig_sag.png|T1 weighted MRI brain in sagittal orientation (provided by the ICBM-MNI152 non linear brain template) | ||
Image:MNI152_lesionload40mL_sag.png|The same MNI152 template with 40mL lesion load simulated lesions (sagittal orientation) | Image:MNI152_lesionload40mL_sag.png|The same MNI152 template with 40mL lesion load simulated lesions (sagittal orientation) | ||
| + | </gallery> | ||
| + | |||
| + | <!-- ---------------------------- --> | ||
| + | {{documentation/{{documentation/version}}/extension-section|Tutorials}} | ||
| + | |||
| + | <gallery> | ||
| + | File:MSLesionSimulator-Tutorial-1.pdf|page=1|How to simulate T1 hypointense Multiple Sclerosis lesions | ||
</gallery> | </gallery> | ||
| Line 60: | Line 65: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|References}} | {{documentation/{{documentation/version}}/extension-section|References}} | ||
| − | * paper | + | * Senra Filho, A. C. da S., Simozo, F. H., dos Santos, A. C., & Junior, L. O. M. (2019). Multiple Sclerosis multimodal lesion simulation tool (MS-MIST). Biomedical Physics & Engineering Express, 5(3), 035003. https://doi.org/10.1088/2057-1976/ab08fc |
| + | |||
| + | Feel free to enter in contact with the authors to request a private copy of the above paper. | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 67: | Line 74: | ||
Repositories: | Repositories: | ||
| − | * Source code: [https://github.com/CSIM-Toolkits/Slicer GitHub repository] | + | * Source code: [https://github.com/CSIM-Toolkits/Slicer-LesionSimulatorExtension GitHub repository] |
| − | * Issue tracker: [https://github.com/CSIM-Toolkits/Slicer/issues open issues and enhancement requests] | + | * Issue tracker: [https://github.com/CSIM-Toolkits/Slicer-LesionSimulatorExtension/issues open issues and enhancement requests] |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-footer}} | {{documentation/{{documentation/version}}/extension-footer}} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 15:44, 9 November 2024
Home < Documentation < Nightly < Extensions < LesionSimulator
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work was partially funded by CAPES and CNPq, a Brazillian Agencies. Information on CAPES can be obtained on the CAPES website and CNPq website. | |||||||||
|
Extension Description
This extension offer a set of tools for brain lesion simulation, based on MRI images. At moment, the module MS Lesion Simulator is available, where it can simulates both baseline scan lesion volumes (given a lesion load) and longitudinal image simulations. In summary, a statistical lesion database is generated based on a set of manual lesion mark-ups, being non-linearly registered to MNI152 space (isotropic 1mm of voxel resolution), and a local contrast ratio is applied on each image modality provided by the user. Using a small set of parameters (lesion load, lesion homogeneity, lesion intensity indenpendence and lesion variability), it is possible to generate a broad range of MS lesions patterns in multimodal MRI imaging techniques (at moment, T1, T2, T2-FLAIR, PD, DTI-FA and DTI-ADC images are provided). For more details about this project, please see the original paper.
Modules
Use Cases
Most frequently used for these scenarios:
- Use Case 1: Simulate different anatomical/clinical MS lesions patterns on healthy individuals MRI images
- In the baseline scan approach, it is offered a simulation procedure where a determined lesion load is reconstructed using a subject specific anatomical features, resulting in a realistic MS lesion load simulation.
- Use Case 2: Simulate longitudinal MS lesion progression on clinical healthy individuals MRI images
- Another important issue in Multiple Sclerosis diagnosis is the lesion progression, where drive the therapeutic strategy by the health professionals. In this case, an automatic segmentation approach could be optimally adjusted for the time progression presented in a exam series.
Tutorials
Similar Extensions
N/A
References
- Senra Filho, A. C. da S., Simozo, F. H., dos Santos, A. C., & Junior, L. O. M. (2019). Multiple Sclerosis multimodal lesion simulation tool (MS-MIST). Biomedical Physics & Engineering Express, 5(3), 035003. https://doi.org/10.1088/2057-1976/ab08fc
Feel free to enter in contact with the authors to request a private copy of the above paper.
Information for Developers
| Section under construction. |
Repositories:
- Source code: GitHub repository
- Issue tracker: open issues and enhancement requests