Difference between revisions of "Slicer3:VisualBlog"
| (94 intermediate revisions by 15 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| − | + | <gallery caption="2010" widths="200px" perrow="4"> | |
| − | <gallery caption="2008" widths="200px" | + | Image:Gimias-running-slicer-module-2010-10-20.png | A set of slicer command line modules are now shipped as part of the [http://www.gimias.org Gimias] tool from UPF in Barcelona! |
| + | Image:800px-NDI booth Slicer MICCAI2010.jpg | Northern Digital, makers of tracking systems widely used in image guided therapy, used Slicer and OpenIGTLink in their booth installation at [http://www.miccai2010.org/ MICCAI2010 in Beijing]. | ||
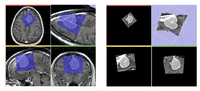
| + | Image:prostate_multiparametric_visualization.png | In this screenshot [[Modules:MainApplicationGUI-Documentation-3.6#Selecting_Among_Standard_Layouts|CompareView layout of 3D Slicer]] is used to facilitate visualization of multiparametric MRI of the prostate | ||
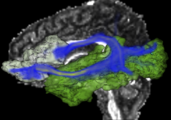
| + | Image:vervet_atlas_template.png| [http://www.nitrc.org/projects/vervet_atlas Vervet Probabilistic Atlas] constructed using 3D Slicer tools is available for download from NITRC. | ||
| + | Image:Avf 3d voronoi big.png| A Voronoi diagram used for [[Modules:VMTKCenterlines|centerline computation]] of a vascular tree. | ||
| + | Image:Vmtkaftercenterline.png| [[Modules:VMTKEasyLevelSetSegmentation|Level-set segmentation]] was used to segment this right coronary artery. | ||
| + | Image:Slicer3.6MeasurementsRulersModel.jpg| [[Modules:Measurements-Documentation-3.6|Rulers and angle widgets]] are available for measuring distances and angles in the 3D scene | ||
| + | Image:deformable_registration_Screenshot1.png| Non-rigid registration module in 3D Slicer can be used to fuse pre-procedural MR images and intra-procedural CT images during cryoablation of renal tumors. The non-rigid registrations are more accurate than rigid registrations. | ||
| + | Image:FourDImage_Screenshot1.png|[[Modules:FourDImage-Documentation-3.6|The 4D Image module]] allows handling a time-series of 3D volume images (4D image), e.g. fMRI, cardiac images, dynamic contrast-enhanced (DCE) images in 3D Slicer. | ||
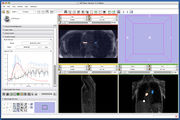
| + | Image:FourDAnalysisModuleScreenShot-3.5.png|Screenshot of [[Modules:FourDAnalysis-Documentation-3.6|4D Analysis Module]] for lung perfusion data. The module provides functionality to analyze time-intensity curves in specified regions. | ||
| + | Image:LipsMuscles.png|Three-dimensional appearance of the lip muscles with three-dimensional isotropic MRI: An in vivo study. R. Olszewski, Y. Liu, T. Duprez, T.M. Xu, H. Reychler: Int J Comput Assist Radiol Surg. 2009 Jun;4(4):349-52. [http://www.slicer.org/slicerWeb/images/4/45/Olszewski-IJCARS2009.pdf See here for the pdf] | ||
| + | Image:RegistrationUsability.png|[[Slicer3:Registration#Registration_in_3D_Slicer|Usability of registration algorithms]]: A joint effort between NA-MIC and NAC is aimed at making advanced registration technology easier to use through better documentation, a solution library and a slicer based solution service. | ||
| + | Image:ScalarBar.png|[[Modules:Colors-Documentation-3.6|A scalar bar]] is available from the colors module | ||
| + | Image:LesionsWithVentricles.png|[[Modules:LesionSegmentationApplications-Documentation-3.6|White matter lesion segmentation]] is now available as an extension to Slicer | ||
| + | Image:RssVentricle.png|[[Modules:RobustStatisticsSeg-Documentation-3.6|RSS]] a new interactive segmentation algorithm | ||
| + | Image:GPUVolumeRender1.png|With [http://vtk.org VTK version 5.6] blazing fast new hardware accelerated volume rendering is available in slicer nightly builds (require nVidia drivers, available on Windows and Linux only). This renderer will be available in Slicer 3.6. | ||
| + | Image:Fastmarching-result-adjusted.jpg|[[Modules:FastMarchingSegmentation-Documentation-3.6|FastMarching segmentation module]] in Slicer 3.6: Segmentation of a meningioma | ||
| + | Image:Corticalthickness.png| Result from the [[Modules:ARCTIC-Documentation-3.6|ARCTIC module]] in Slicer 3.6: cortical thickness map from pediatric MRI | ||
| + | Image:SynchroGrab4D titleShot.jpg|[http://hdl.handle.net/10380/3083 SynchroGrab4D] reconstructs 4D (3D+time) ultrasound datasets in real-time, using a tracked 2D transducer. It is particularly useful for intraoperative imaging of moving organs, including the heart and those influenced by respiratory motion, such as the liver. | ||
| + | Image:ExtractSubvolumeROI_gallery.png|[[Modules:CropVolume-Documentation-3.6|CropVolume is a new interactive GUI module in Slicer 3.6]] that allows to extract and resample an arbitrary parallelepiped shaped region from a larger scalar image volume. | ||
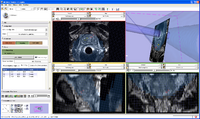
| + | Image:ProstateNav_gallery.png|[[Modules:ProstateNav-Documentation-3.6|ProstateNav]] is an interactive GUI module in Slicer 3.6 for MRI guided robot assisted biopsy of the prostate. It supports multiple needle positioning devices: transrectal and transperineal robot and transperineal template. | ||
| + | Image:RegistrationResults2.jpg| [[Modules:DeformableB-SplineRegistration-Documentation-3.6|B-Spline Registration]] in Slicer 3.6 allows for non-rigid registration between pre-procedure MRI and intra-procedure CT images during CT guided tumor ablation in the liver. It provides increased tumor visualization during the planning, targeting and monitoring phases of the ablation procedure. | ||
| + | Image:IntraOpDTI.jpg| Intra-operative querying of white matter tracts is now possible by interfacing Slicer 3.6 to the commercial [http://www.brainlab.com/scripts/website_english.asp BrainLab] system and Yale's [http://bioimagesuite.org/ BioImageSuite] software. | ||
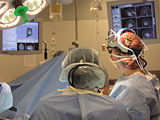
| + | Image:BetaProbe.jpg| Intra-operative navigation of a positron probe can help detect regions of high [18F]-FDG uptake, potentially improving tumor removal at resection borders. The first two image shows a PET/CT scan of the phantom showing where radiation is located, and the third image shows results as indicated by the positron probe. | ||
| + | </gallery> | ||
| + | <gallery caption="2009" widths="200px" perrow="4"> | ||
| + | Image:Konnectivity-2009-12-23.png|A frame from [http://aokhok.unideb.hu/konnektivity/ Andras Jakab's interactively zoomable super high-res tractography image!] | ||
| + | Image:Qt-CLI-2009-11-29.png|Frame from a movie showing Qt Command Line Modules. See whole movie [http://www.youtube.com/watch?v=e_T2cYGZmSw on youtube]. | ||
| + | Image:Qt-KWW-Transforms-2009-11-24.png|Frame from a movie showing Qt Transforms module. See whole movie [http://www.youtube.com/watch?v=oJFSJWBUFEI on youtube] or download [[file:QSlicerTransformsModule.ogg]]. | ||
| + | Image:VolumeRenderingICPE.png|Volume rendering: Illustrative Context Preserving Exploration | ||
| + | Image:Dual 3D View.png|Multiple 3D rendering windows | ||
| + | Image:QMRMLWidgetPlugins-Screenshot.png|Early results of the [[Qt_in_Slicer3]] effort showing new QMRMLWidgets as a plug-in for the QtDesigner application. | ||
| + | Image:Nagy-simicircular-canals-2009.png|Attila Nagy and his colleagues created a [http://www.orl.szote.u-szeged.hu/~attila/presentations.html very nice set of presentations and animations] with slicer to illustrate 3D reconstruction as applied to otorhinolaryngology. (See [[Nagy-videos-2009:Notes|Notes]]). | ||
| + | Image:Slicer3-mac-64bit-2009-09-11.mov|Interactive volume rendering of 512x512x1749 CT volume on 64 bit build of slicer for Mac OSX 10.6. | ||
| + | Image:Registration-module-hierarchy-prototype.png|Proptype hierarchy of registration modules for specific use-cases that present a simpler view of the capabilities of RegisterImages. | ||
| + | Image:Xoran-Cone-beam-CT_Craniofacial_Asymetry-2009-08-27.png|Example experiment in craniofacial surgery planning by mirroring one side of the skull to the other. This example was done in collaboration with Will van Kampen of [http://xorantech.com Xoran Technologies] and was a follow up to the August 25 [http://www.na-mic.org/Wiki/index.php/Events:Slicer_Workshop_August_2009 Slicer IGT Workshop] in Boston. | ||
| + | Image:Bioimagesuite slicer1.png|[http://www.bioimagesuite.org BioImageSuite] module being discovered. | ||
| + | Image:Bioimagesuite slicer2.png|[http://www.bioimagesuite.org BioImageSuite] modules in menu. | ||
| + | Image:Bioimagesuite slicer3.png|Autogenerated [http://www.bioimagesuite.org BioImageSuite] module GUI. | ||
| + | Image:Bioimagesuite slicer4.png|Slicer command line module recognized and loaded in [http://www.bioimagesuite.org BioImageSuite]. | ||
| + | Image:BIS-Slicer-2009-08-13.jpg|Xenios Papademetris enabled the tools in [http://www.bioimagesuite.org BioImageSuite] to work as command line modules in slicer. | ||
| + | Image:2009JuneJulyStats.png|See [http://www.slicer.org/pages/Special:Download_Stats?order=downloads&reverse=1 here] for download statistics (page might take a while to load). The Piechart demonstrates the download proportions between the different OS. | ||
| + | Image:Stochastic-Tractography-01040-lh-all-3D-cropped.png|[http://www.na-mic.org/Wiki/index.php/Summer2009:VCFS '''New Stochastic Tractography'''] tools for analysis of diffusion volumes implemented as Python modules by Julien de Siebenthal. | ||
| + | Image:3DSlicerFourDAnalysis Screenshot.png |[http://www.na-mic.org/Wiki/index.php/2009_Summer_Project_Week_4D_Imaging '''Four Dimensional Image (Time Series) Analysis'''] Junichi Tokuda has implemented a slicer module for dynamic MR analysis and a general framework for working with time series volume groups. | ||
| + | Image:Slicer dti seeding or.jpg|[http://www.na-mic.org/Wiki/index.php/2009_Summer_Project_Week_Slicer3_Brainlab_Introduction '''Intraoperative navigation experiments'''] integrate slicer3 with the commercial [http://www.brainlab.com/scripts/website_english.asp BrainLab] system and Yale's [http://bioimagesuite.org/ BioImageSuite] software. | ||
| + | Image:SkullStrippingInSlicer.png|'''New [http://www.na-mic.org/Wiki/index.php/2009_Summer_Project_Week_Skull_Stripping skull stripping extension module]''' implemented by Xiaodong Tao. | ||
| + | Image:MS-TSA-CompareView.png|'''CompareView for Longitudinal Visualization of MS''' implemented by Jim Miller and demonstrated using a set of 5 scans of a patient with multiple sclerosis. Data provided by Dominik Meier of the [http://cni.bwh.harvard.edu Center for Neurological Imaging] | ||
| + | </gallery> | ||
| + | <gallery caption="2008" widths="200px" perrow="4"> | ||
| + | Image:Synarc-Li-Level tracing.gif|'''Constrained Level Tracing Algorithm''' implemented by Andrew Li and his team at [http://synarc.com/ Synarc] built on the Slicer3 Editor module has been deployed for bone density measurements in support of clinical trials. Synarc employees participated in the [http://www.na-mic.org/Wiki/index.php/Stanford_2008_Slicer_Workshop 2008 Slicer Training] event at Stanford University. | ||
| + | Image:DebrecenTumorMovieStartFrame.png|'''Start frame from [[Tumor_Movie_from_Debrecen|time lapse movie]] showing "Chameleon Tumor" progression.''' Video courtesy by Ervin Berenyi and Andras Jakab, Department of Medical Laboratory and Diagnostic Imaging, University of Debrecen Medical School and Health Science Center. | ||
| + | Image:Femesh-in-trunk-120808.png|'''Image showing the [http://www.na-mic.org/Wiki/index.php/NA-MIC_NCBC_Collaboration:Automated_FE_Mesh_Development IA-FEMesh] module in Slicer3.''' <br> This image shows the display of material properties assigned to a hexahedral mesh. This image was created by Curl Lisle, Nicole Grosland, Kiran Shivanna, Steve Pieper, and Vincent Magnotta | ||
| + | Image:VascularLesionSegmentation.jpg|'''Image showing the segmentation of vasular lesions based on T1, T2 and FLAIR images.'''<br>A Bayesian classification is performed to label each of the voxels as grey matter, white matter, CSF, and lesion. | ||
| + | Image:ClipModel-with-widget.png|The new Clip Model Module (Alex Yarmarkovich) uses a VTK box widget to select cutting planes to clip a selected model. | ||
| + | Image:PythonDistanceTransform.png|Distance Transform from a label map calculated by Steve Pieper using the NumpyScript module created by Luca Antiga. See [[Slicer3:Python]] for more info. | ||
| + | Image:DTI-Visualization-Neurosurgical-Tutorial.png|Exploring DTI visualization using the [http://wiki.na-mic.org/Wiki/index.php/Slicer3.2:Training Neurosurgical Tutorial]. A virtual probe (fiducial) was positioned in the motor cortex and used as a seed for tractography. The region adjacent to the tumor was used as a seeding area for a separate tractography, which is represented as thin lines with ellipsoid glyphs. | ||
| + | Image:Color-image-reslice-2008-06-11.png|'''Color Image Reslice''' Demonstrates the ability to read a sequence of jpg files as a color volume and apply 3D interactive reslice tools. Image data is Visible Human 2 courtesy of the [http://nac.spl.harvard.edu/pages/Research_Cores#Clinical_Computational_Anatomy_Core NAC Clinical Computational Anatomy Core] | ||
| + | Image:Andras-Jakab-Debrecen-gamma01.jpg|'''Planning for CT Radiosurgery augmented with diffusion tractography (fibers: internal capsule, corpus callosum and the environment of the tumor, which is a metastatic tumor in the parietal lobe)''' Image Courtesy of University of Debrecen, Medical School and Health Science Center [[Debrecen | more information...]] | ||
| + | Image:Debrecen slicer 01.jpg|'''Integration of diffusion and structural imaging for tumor visualization''' Image Courtesy of University of Debrecen, Medical School and Health Science Center | ||
Image:Slicer_IGTL_NITRobot.jpg|'''Integration of Neurosurgical Robot and Slicer using OpenIGTLink, February 2008'''<br>Joint team of Nagoya Institute of Technology and Brigham and Women's Hospital demonstrated a prototype integration of neurosurgical robot and 3D Slicer using OpenIGTLink. | Image:Slicer_IGTL_NITRobot.jpg|'''Integration of Neurosurgical Robot and Slicer using OpenIGTLink, February 2008'''<br>Joint team of Nagoya Institute of Technology and Brigham and Women's Hospital demonstrated a prototype integration of neurosurgical robot and 3D Slicer using OpenIGTLink. | ||
Image:Cardioseg+volume.png|'''Cardiac segmentation and CT Volume Rendering, February 2008'''<br> Using data and segmentations from the [http://wiki.na-mic.org/Wiki/index.php/NA-MIC_Childrens_Collaboration collaboration with Boston Children's Hospital Pediatric Cardiology]. | Image:Cardioseg+volume.png|'''Cardiac segmentation and CT Volume Rendering, February 2008'''<br> Using data and segmentations from the [http://wiki.na-mic.org/Wiki/index.php/NA-MIC_Childrens_Collaboration collaboration with Boston Children's Hospital Pediatric Cardiology]. | ||
Image:Slicer_IGTL_PartialImage.png|'''Partial image update using OpenIGTLink, February 2008'''<br> [http://wiki.na-mic.org/Wiki/index.php/OpenIGTLink OpenIGTLink] allows an external imaging scanner (Ultrasound/CT/MR) to update part of a volume, which has already been loaded on the Slicer. This is helpful if the imaging plane sweeps the subject, and images are transfered to the Slicer on-the-fly. | Image:Slicer_IGTL_PartialImage.png|'''Partial image update using OpenIGTLink, February 2008'''<br> [http://wiki.na-mic.org/Wiki/index.php/OpenIGTLink OpenIGTLink] allows an external imaging scanner (Ultrasound/CT/MR) to update part of a volume, which has already been loaded on the Slicer. This is helpful if the imaging plane sweeps the subject, and images are transfered to the Slicer on-the-fly. | ||
| − | Image:Slicer_CudaHead.jpg|'''The First MRML node rendering using CUDA February 2008'''<br> [ | + | Image:Slicer_CudaHead.jpg|'''The First MRML node rendering using CUDA February 2008'''<br> [[Slicer3:Volume_Rendering_With_Cuda| Cuda Volume Rendering]] provides a Volume Rendering Method on the new and advanced NVidia Hardware. |
Image:Slicer_OpenIGTLINK.png|'''The First Implementation of OpenIGTLink, January 2008'''<br> [http://wiki.na-mic.org/Wiki/index.php/OpenIGTLink OpenIGTLink] protocol provides plug-and-play connectivity to tracking devices, imagers (MR, ultrasound ...) and other medical devices. A real-time image transfer to the Slicer in 10 fps is demonstrated in the screenshot. | Image:Slicer_OpenIGTLINK.png|'''The First Implementation of OpenIGTLink, January 2008'''<br> [http://wiki.na-mic.org/Wiki/index.php/OpenIGTLink OpenIGTLink] protocol provides plug-and-play connectivity to tracking devices, imagers (MR, ultrasound ...) and other medical devices. A real-time image transfer to the Slicer in 10 fps is demonstrated in the screenshot. | ||
Image:Slicer_TrackerDaemon_IGSTK.png|'''Image from Haiying Liu, Patrick Cheng, Noby Hata, Junichi Tokuda, Luis Ibanez, and Steve Pieper, January 2008.''' <br>In the NAMIC All Hands Meeting 2008, the bi-directional socket communication has been established between the Tracker Daemon in Slicer 3 and IGSTK server which acquires tracking data from device and sends it to Tracker Daemon. The speed of communication is controlled by Slicer. | Image:Slicer_TrackerDaemon_IGSTK.png|'''Image from Haiying Liu, Patrick Cheng, Noby Hata, Junichi Tokuda, Luis Ibanez, and Steve Pieper, January 2008.''' <br>In the NAMIC All Hands Meeting 2008, the bi-directional socket communication has been established between the Tracker Daemon in Slicer 3 and IGSTK server which acquires tracking data from device and sends it to Tracker Daemon. The speed of communication is controlled by Slicer. | ||
</gallery> | </gallery> | ||
| − | <gallery caption="2007" widths="200px" | + | <gallery caption="2007" widths="200px" perrow="4"> |
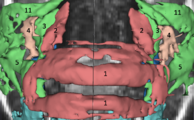
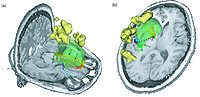
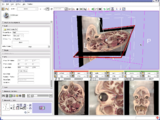
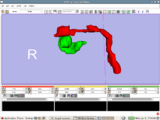
Image:ENT.png|'''Image from Jaesung Hong December 2007'''<br> Visualization of cochlea(green) and facial nerve(red) for ENT navigation. We are moving on from Slicer2 to Slicer3 at Kyushu University Hospital in Japan. | Image:ENT.png|'''Image from Jaesung Hong December 2007'''<br> Visualization of cochlea(green) and facial nerve(red) for ENT navigation. We are moving on from Slicer2 to Slicer3 at Kyushu University Hospital in Japan. | ||
Image:Slicer-igstk.png|'''Image from Steve Pieper, Luis Ibanez, Haiying Liu December 2007'''<br>Result of [http://wiki.na-mic.org/Wiki/index.php/2007_December_Slicer_IGT_Programming Slicer for IGT workshop] showing Slicer3 transform node being updated by a [http://www.igstk.org IGSTK] tracker process. | Image:Slicer-igstk.png|'''Image from Steve Pieper, Luis Ibanez, Haiying Liu December 2007'''<br>Result of [http://wiki.na-mic.org/Wiki/index.php/2007_December_Slicer_IGT_Programming Slicer for IGT workshop] showing Slicer3 transform node being updated by a [http://www.igstk.org IGSTK] tracker process. | ||
| Line 18: | Line 76: | ||
Image:sirp1.png|'''Image from wjp on Wednesday, October 24, 2007'''<br>QueryAtlas visualizing combined morphology and functional analyses generated by running FreeSurfer and a FIPS pipeline on a BIRN phaseII SIRP dataset. Interactive annotations (on mouse-over) are being translated thru Slicer's controlled vocabulary. | Image:sirp1.png|'''Image from wjp on Wednesday, October 24, 2007'''<br>QueryAtlas visualizing combined morphology and functional analyses generated by running FreeSurfer and a FIPS pipeline on a BIRN phaseII SIRP dataset. Interactive annotations (on mouse-over) are being translated thru Slicer's controlled vocabulary. | ||
Image:SlicerReformat.png|'''Image from Jim on Saturday, September 15, 2007'''<br>Slice viewers can be used to specify oblique reformats using the 'Reformat' orientation (instead of axial, coronal, sagittal) and CTRL-Right-Button-Move<br>(subject to change). | Image:SlicerReformat.png|'''Image from Jim on Saturday, September 15, 2007'''<br>Slice viewers can be used to specify oblique reformats using the 'Reformat' orientation (instead of axial, coronal, sagittal) and CTRL-Right-Button-Move<br>(subject to change). | ||
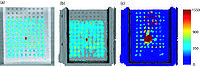
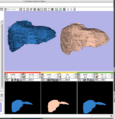
| − | Image:Livers.png|'''Image from Dirk, Matthew, Jim, and Steve on Monday, August 13, 2007'''<br>A new label map smoothing tool has been added to help with our [ | + | Image:Livers.png|'''Image from Dirk, Matthew, Jim, and Steve on Monday, August 13, 2007'''<br>A new label map smoothing tool has been added to help with our [http://wiki.na-mic.org/Wiki/index.php/NA-MIC_Collaborations#Children.27s_Pediatric_Cardiology_Collaboration_with_SCI.2FSPL.2FNortheastern collaboration with Children's Hospital Boston, SCI at University of Utah and Northeastern University]. The unfiltered labelmap is shown in blue, and the filtered results are shown in peach. |
Image:Editbox.png|'''Image from Steve and Wendy on Monday, August 6, 2007'''<br>New Editor functionality, with EditBox which is invoked using the F1 key (will soon be moved to the space bar). | Image:Editbox.png|'''Image from Steve and Wendy on Monday, August 6, 2007'''<br>New Editor functionality, with EditBox which is invoked using the F1 key (will soon be moved to the space bar). | ||
| − | Image:EMSegment31Structures.png|'''Image from Brad and Kilian on Wednesday, June, 21, 2007'''<br>Example of [ | + | Image:EMSegment31Structures.png|'''Image from Brad and Kilian on Wednesday, June, 21, 2007'''<br>Example of [http://wiki.na-mic.org/Wiki/index.php/Projects/Structural/2007_Project_Week_EMSegmentation_Validation EMSegmenter in Slicer3] |
| − | Image:ConnectivityMap.png|'''Image from pieper on Friday, June, 9, 2007'''<br>Slicer3 Module for [ | + | Image:ConnectivityMap.png|'''Image from pieper on Friday, June, 9, 2007'''<br>Slicer3 Module for [http://wiki.na-mic.org/Wiki/index.php/Algorithm:MIT:DTI_StochasticTractography Stochastic Tractography] from MIT (Ngo, Golland) and BWH (Westin, Kubicki). |
Image:kwmeshvisu-slicer-logo.png|'''Image from ipek on Wednesday, June, 6, 2007'''<br>UNC Logo in Slicer3 for KWMeshVisu (Ipek Oguz, Martin Styner). | Image:kwmeshvisu-slicer-logo.png|'''Image from ipek on Wednesday, June, 6, 2007'''<br>UNC Logo in Slicer3 for KWMeshVisu (Ipek Oguz, Martin Styner). | ||
Image:CineDisplayDesigns.png|'''Image from wjp on Wednesday, May, 30, 2007'''<br>Design mockups for Slicer3's Cine Display interface (William Leue, Wendy Plesniak). | Image:CineDisplayDesigns.png|'''Image from wjp on Wednesday, May, 30, 2007'''<br>Design mockups for Slicer3's Cine Display interface (William Leue, Wendy Plesniak). | ||
| Line 27: | Line 85: | ||
Image:EMSegmentation_Results_Screenshot.png|'''Image from davisb on Monday, April, 27, 2007'''<br>EMSegment screenshot---segmentation results and work-flow GUI | Image:EMSegmentation_Results_Screenshot.png|'''Image from davisb on Monday, April, 27, 2007'''<br>EMSegment screenshot---segmentation results and work-flow GUI | ||
Image:Mimx.png|'''Image from magnotta on Monday, April, 16, 2007'''<br>Mimx Logo in Slicer3 for VoxelMeshingModule | Image:Mimx.png|'''Image from magnotta on Monday, April, 16, 2007'''<br>Mimx Logo in Slicer3 for VoxelMeshingModule | ||
| − | Image:PythonMenu.png |'''Image from blezek on Tuesday, April, 10, 2007 at 8:00AM'''<br>Python incorporated into [[Slicer3:Python | Slicer]]. [[Slicer3:VisualBlog_Extension# | + | Image:PythonMenu.png |'''Image from blezek on Tuesday, April, 10, 2007 at 8:00AM'''<br>Python incorporated into [[Slicer3:Python| Slicer]]. [[Slicer3:VisualBlog_Extension#Image_from_blezek_on_Tuesday.2C_April.2C_10.2C_2007_at_8:00AM | More ...]] |
Image:ZoomWindow.png|'''Image from wjp on Thursday, March 22, 2007 at 10:00AM'''<br>New GUI elements: Shows a magnified view in the GUI panel of an area around the mouse in any Slice Window. | Image:ZoomWindow.png|'''Image from wjp on Thursday, March 22, 2007 at 10:00AM'''<br>New GUI elements: Shows a magnified view in the GUI panel of an area around the mouse in any Slice Window. | ||
Image:NavigationWindow.png|'''Image from wjp on Thursday, March 22, 2007 at 10:00AM'''<br>New GUI elements: Shows birds-eye-view of the scene relative to the outline of the 3D Viewer's window in the 'Manipulate 3D View' GUI panel. | Image:NavigationWindow.png|'''Image from wjp on Thursday, March 22, 2007 at 10:00AM'''<br>New GUI elements: Shows birds-eye-view of the scene relative to the outline of the 3D Viewer's window in the 'Manipulate 3D View' GUI panel. | ||
Image:Slicer3FirstTractography.png|'''Image from pieper on Tuesday, January 16, 2007 at 7:58PM'''<br>From Lauren, shows fiber tracts loaded from a file and with display properties controlled by the GUI. | Image:Slicer3FirstTractography.png|'''Image from pieper on Tuesday, January 16, 2007 at 7:58PM'''<br>From Lauren, shows fiber tracts loaded from a file and with display properties controlled by the GUI. | ||
</gallery> | </gallery> | ||
| − | <gallery caption="2006" widths="200px" | + | <gallery caption="2006" widths="200px" perrow="4"> |
Image:S3GUI-12-29-06.png|'''Image from wjp on Friday, December 29, 2006 at 3:23PM'''<br>Module choose and navigation functionality integrated into the application toolbar to create more space for module GUIs on the side panel. Slice Controller widgets also updated to have more functionality available, a button to link and unlink their control, and an improved visual design. | Image:S3GUI-12-29-06.png|'''Image from wjp on Friday, December 29, 2006 at 3:23PM'''<br>Module choose and navigation functionality integrated into the application toolbar to create more space for module GUIs on the side panel. Slice Controller widgets also updated to have more functionality available, a button to link and unlink their control, and an improved visual design. | ||
Image:S3GUIWithLogo.png|'''Image from wjp on Monday, November 28, 2006 at 6:40M'''<br>New 3D Slicer logo integrated with slicer GUI. | Image:S3GUIWithLogo.png|'''Image from wjp on Monday, November 28, 2006 at 6:40M'''<br>New 3D Slicer logo integrated with slicer GUI. | ||
Latest revision as of 23:18, 2 July 2011
Home < Slicer3:VisualBlog- 2010
A set of slicer command line modules are now shipped as part of the Gimias tool from UPF in Barcelona!
Northern Digital, makers of tracking systems widely used in image guided therapy, used Slicer and OpenIGTLink in their booth installation at MICCAI2010 in Beijing.
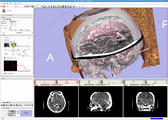
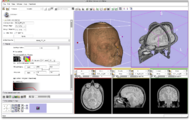
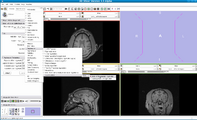
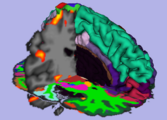
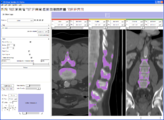
In this screenshot CompareView layout of 3D Slicer is used to facilitate visualization of multiparametric MRI of the prostate
Vervet Probabilistic Atlas constructed using 3D Slicer tools is available for download from NITRC.
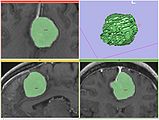
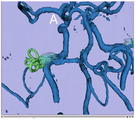
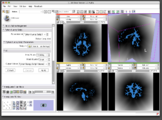
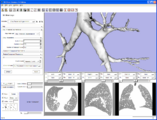
A Voronoi diagram used for centerline computation of a vascular tree.
Level-set segmentation was used to segment this right coronary artery.
Rulers and angle widgets are available for measuring distances and angles in the 3D scene
The 4D Image module allows handling a time-series of 3D volume images (4D image), e.g. fMRI, cardiac images, dynamic contrast-enhanced (DCE) images in 3D Slicer.
Screenshot of 4D Analysis Module for lung perfusion data. The module provides functionality to analyze time-intensity curves in specified regions.
Three-dimensional appearance of the lip muscles with three-dimensional isotropic MRI: An in vivo study. R. Olszewski, Y. Liu, T. Duprez, T.M. Xu, H. Reychler: Int J Comput Assist Radiol Surg. 2009 Jun;4(4):349-52. See here for the pdf
Usability of registration algorithms: A joint effort between NA-MIC and NAC is aimed at making advanced registration technology easier to use through better documentation, a solution library and a slicer based solution service.
A scalar bar is available from the colors module
White matter lesion segmentation is now available as an extension to Slicer
RSS a new interactive segmentation algorithm
With VTK version 5.6 blazing fast new hardware accelerated volume rendering is available in slicer nightly builds (require nVidia drivers, available on Windows and Linux only). This renderer will be available in Slicer 3.6.
FastMarching segmentation module in Slicer 3.6: Segmentation of a meningioma
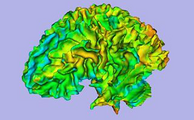
Result from the ARCTIC module in Slicer 3.6: cortical thickness map from pediatric MRI
SynchroGrab4D reconstructs 4D (3D+time) ultrasound datasets in real-time, using a tracked 2D transducer. It is particularly useful for intraoperative imaging of moving organs, including the heart and those influenced by respiratory motion, such as the liver.
CropVolume is a new interactive GUI module in Slicer 3.6 that allows to extract and resample an arbitrary parallelepiped shaped region from a larger scalar image volume.
ProstateNav is an interactive GUI module in Slicer 3.6 for MRI guided robot assisted biopsy of the prostate. It supports multiple needle positioning devices: transrectal and transperineal robot and transperineal template.
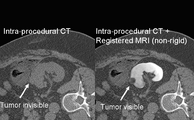
B-Spline Registration in Slicer 3.6 allows for non-rigid registration between pre-procedure MRI and intra-procedure CT images during CT guided tumor ablation in the liver. It provides increased tumor visualization during the planning, targeting and monitoring phases of the ablation procedure.
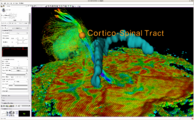
Intra-operative querying of white matter tracts is now possible by interfacing Slicer 3.6 to the commercial BrainLab system and Yale's BioImageSuite software.
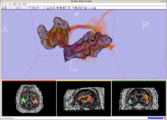
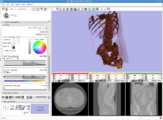
Intra-operative navigation of a positron probe can help detect regions of high [18F]-FDG uptake, potentially improving tumor removal at resection borders. The first two image shows a PET/CT scan of the phantom showing where radiation is located, and the third image shows results as indicated by the positron probe.
- 2009
Frame from a movie showing Qt Command Line Modules. See whole movie on youtube.
Frame from a movie showing Qt Transforms module. See whole movie on youtube or download File:QSlicerTransformsModule.ogg.
Early results of the Qt_in_Slicer3 effort showing new QMRMLWidgets as a plug-in for the QtDesigner application.
Attila Nagy and his colleagues created a very nice set of presentations and animations with slicer to illustrate 3D reconstruction as applied to otorhinolaryngology. (See Notes).
Interactive volume rendering of 512x512x1749 CT volume on 64 bit build of slicer for Mac OSX 10.6.
Example experiment in craniofacial surgery planning by mirroring one side of the skull to the other. This example was done in collaboration with Will van Kampen of Xoran Technologies and was a follow up to the August 25 Slicer IGT Workshop in Boston.
BioImageSuite module being discovered.
BioImageSuite modules in menu.
Autogenerated BioImageSuite module GUI.
Slicer command line module recognized and loaded in BioImageSuite.
Xenios Papademetris enabled the tools in BioImageSuite to work as command line modules in slicer.
See here for download statistics (page might take a while to load). The Piechart demonstrates the download proportions between the different OS.
New Stochastic Tractography tools for analysis of diffusion volumes implemented as Python modules by Julien de Siebenthal.
Four Dimensional Image (Time Series) Analysis Junichi Tokuda has implemented a slicer module for dynamic MR analysis and a general framework for working with time series volume groups.
Intraoperative navigation experiments integrate slicer3 with the commercial BrainLab system and Yale's BioImageSuite software.
New skull stripping extension module implemented by Xiaodong Tao.
CompareView for Longitudinal Visualization of MS implemented by Jim Miller and demonstrated using a set of 5 scans of a patient with multiple sclerosis. Data provided by Dominik Meier of the Center for Neurological Imaging
- 2008
Constrained Level Tracing Algorithm implemented by Andrew Li and his team at Synarc built on the Slicer3 Editor module has been deployed for bone density measurements in support of clinical trials. Synarc employees participated in the 2008 Slicer Training event at Stanford University.
Start frame from time lapse movie showing "Chameleon Tumor" progression. Video courtesy by Ervin Berenyi and Andras Jakab, Department of Medical Laboratory and Diagnostic Imaging, University of Debrecen Medical School and Health Science Center.
Image showing the IA-FEMesh module in Slicer3.
This image shows the display of material properties assigned to a hexahedral mesh. This image was created by Curl Lisle, Nicole Grosland, Kiran Shivanna, Steve Pieper, and Vincent MagnottaDistance Transform from a label map calculated by Steve Pieper using the NumpyScript module created by Luca Antiga. See Slicer3:Python for more info.
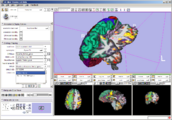
Exploring DTI visualization using the Neurosurgical Tutorial. A virtual probe (fiducial) was positioned in the motor cortex and used as a seed for tractography. The region adjacent to the tumor was used as a seeding area for a separate tractography, which is represented as thin lines with ellipsoid glyphs.
Color Image Reslice Demonstrates the ability to read a sequence of jpg files as a color volume and apply 3D interactive reslice tools. Image data is Visible Human 2 courtesy of the NAC Clinical Computational Anatomy Core
Planning for CT Radiosurgery augmented with diffusion tractography (fibers: internal capsule, corpus callosum and the environment of the tumor, which is a metastatic tumor in the parietal lobe) Image Courtesy of University of Debrecen, Medical School and Health Science Center more information...
Cardiac segmentation and CT Volume Rendering, February 2008
Using data and segmentations from the collaboration with Boston Children's Hospital Pediatric Cardiology.Partial image update using OpenIGTLink, February 2008
OpenIGTLink allows an external imaging scanner (Ultrasound/CT/MR) to update part of a volume, which has already been loaded on the Slicer. This is helpful if the imaging plane sweeps the subject, and images are transfered to the Slicer on-the-fly.The First MRML node rendering using CUDA February 2008
Cuda Volume Rendering provides a Volume Rendering Method on the new and advanced NVidia Hardware.The First Implementation of OpenIGTLink, January 2008
OpenIGTLink protocol provides plug-and-play connectivity to tracking devices, imagers (MR, ultrasound ...) and other medical devices. A real-time image transfer to the Slicer in 10 fps is demonstrated in the screenshot.Image from Haiying Liu, Patrick Cheng, Noby Hata, Junichi Tokuda, Luis Ibanez, and Steve Pieper, January 2008.
In the NAMIC All Hands Meeting 2008, the bi-directional socket communication has been established between the Tracker Daemon in Slicer 3 and IGSTK server which acquires tracking data from device and sends it to Tracker Daemon. The speed of communication is controlled by Slicer.
- 2007
Image from Steve Pieper, Luis Ibanez, Haiying Liu December 2007
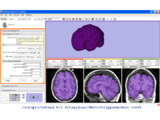
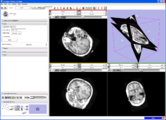
Result of Slicer for IGT workshop showing Slicer3 transform node being updated by a IGSTK tracker process.Image from Steve Pieper, December 2007
Display of segmented heart data results using volume rendering inside Slicer3. See JHU Computational Anatomy Portal for more information.Image from Tri Ngo and Steve Pieper, November 2007
Display of Stochastic Tractography results using volume rendering inside Slicer3. See here for more information.Image from Andy Freudling on October 2007
First results of volume rendering inside Slicer3. See here for more information.Image from Dirk, Matthew, Jim, and Steve on Monday, August 13, 2007
A new label map smoothing tool has been added to help with our collaboration with Children's Hospital Boston, SCI at University of Utah and Northeastern University. The unfiltered labelmap is shown in blue, and the filtered results are shown in peach.Image from Brad and Kilian on Wednesday, June, 21, 2007
Example of EMSegmenter in Slicer3Image from pieper on Friday, June, 9, 2007
Slicer3 Module for Stochastic Tractography from MIT (Ngo, Golland) and BWH (Westin, Kubicki).
- 2006
Image from wjp on Friday, December 29, 2006 at 3:23PM
Module choose and navigation functionality integrated into the application toolbar to create more space for module GUIs on the side panel. Slice Controller widgets also updated to have more functionality available, a button to link and unlink their control, and an improved visual design.Image from pieper on Tuesday, August 08, 2006 at 4:26PM
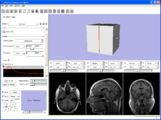
A number of things have come together to allow the new prototype editor module. More ...Image from pieper on Thursday, July 27, 2006 at 5:21PM
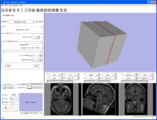
Shows that a single pixel voxel in black is exactly bounded by a unit cube in RAS space. Also the blinking eye buttons for slice visiblility are hooked up and there is now a red-yellow-green color coding for the slice windows (carried over from slicer2).Image from pieper on Thursday, July 13, 2006 at 4:49PM
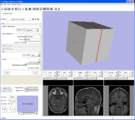
Test of interactive slice textured plane control. Not yet eneabled through GUI, but driven by a test script that you can source into the console. More ...Image from pieper on Monday, July 03, 2006 at 3:35PM
Test of the new logo widget for adding module-specific watermark logos directly in the 3d view. Uses a vtkLogoWidget from VTK cvs head. Kitware is working on cmake support to notify developers when a VTK cvs update is needed. Once that is done, the logo code will be added to slicer3 svn.
About the VisualBlog
The VisualBlog is meant to be an easy place to upload screenshot so that both developers and outside observes can track the progress of the project.
Many thanks to developers and users for contributing their images here.