Difference between revisions of "Slicer4:VisualBlog"
m (Text replacement - "\[https:\/\/www\.slicer\.org\/slicerWiki\/index.php\/([^ ]+) ([^]]+)]" to "$2") |
|||
| (59 intermediate revisions by 16 users not shown) | |||
| Line 1: | Line 1: | ||
| + | This is the Slicer 4 visual blog. Please click [[Slicer3:VisualBlog| here]] if you are looking for the old Slicer 3 visual blog. | ||
| + | |||
| + | <gallery caption="2016" widths="200px" perrow="4"> | ||
| + | Image:2016-SlicerBlog Lumpectomy.png | Clinical research: 3D Slicer is used in real-time navigation of breast cancer surgery at Queen's University, Kingston, ON, Canada. The method is published in the following paper: [http://dx.doi.org/10.1109/TBME.2015.2466591 Ungi T, Gauvin G, Lasso A, Yeo CT, Pezeshki P, Vaughan T, Carter K, Rudan J, Engel CJ, Fichtinger G. Navigated Breast Tumor Excision Using Electromagnetically Tracked Ultrasound and Surgical Instruments. IEEE Trans Biomed Eng. 2016 Mar;63(3):600-6.]. | ||
| + | </gallery> | ||
| + | |||
| + | <gallery caption="2014" widths="200px" perrow="4"> | ||
| + | Image:2014-08-26-SlicerIGT-EMBC2014Tutorial.png | Pre-conference tutorial session [http://www.slicerigt.org/wp/embc-2014-tutorial/ Building image-guidance systems from open-source components] at IEEE EMBC in Chicago (August 26, 2014). | ||
| + | Image:2014-06-27-SlicerKuka.png|Demo at CARS 2014 in Fukuoka, Japan. Slicer is connected to a KUKA robot for visualization of 3D models of the robot, the anatomy, and the workspace. The system is originally developed at [http://www.na-mic.org/Wiki/index.php/2013_Summer_Project_Week:kukarobot NA-MIC Summer Project Week]. | ||
| + | Image:2014AppleVisualization.png| Multiplanar and volume rendered view of a [http://forge.abcd.harvard.edu/gf/download/frsrelease/85/2945/GrannySmith.zip CT scan of a Granny Smith apple]. In plane resolution is 0.33 mm, and 1.25 mm slice thickness. | ||
| + | Image:Post ant R oblique clean.JPG | Render created in [http://blender.org Blender] of data segmented and exported from Slicer. [http://ivaladesign.com/ Taimur Alavi] created this from a CT scan of a friend who had broken his neck. | ||
| + | Image:IMG 20140213 101529.jpg | 3D Print of the same broken neck model, given to the friend as a kind of "good luck charm". The friend recovered. | ||
| + | Image:TCIABrowserLogo.png|The [[Documentation/Nightly/Extensions/TCIABrowser|TCIABrowser extension]] TCIABrowser gives you access to 18 collections of The Cancer Imaging Archive (http://cancerimagingarchive.net/), which contains a total of over 23,000 image series from a variety of manufacturers and of different modalities and organs. | ||
| + | </gallery> | ||
| + | |||
| + | <gallery caption="2013" widths="200px" perrow="4"> | ||
| + | Image:ShapeModelSequenceBrowsing3dOnly.gif| The [[Documentation/Nightly/Extensions/MultidimData|Multidimensional data extension]] allows creating, editing, and visualizing multidimensional data sets in 3D Slicer. Browsing, replaying 4D volumes, reviewing deforming 3D models, replaying tracked ultrasound, tool navigation data in 3D, applying time-varying transforms are now all possible. | ||
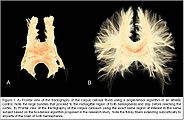
| + | Image:UKFTractography_CC.jpg|[[Documentation/Nightly/Extensions/UKFTractography|UKFTractography]] is a module for computing tractography of DWI images using an unscented Kalman filter. Because of its 2-tensor algorithm, it is able to model fiber crossings and capture many more fibers than a single tensor algorithm. | ||
| + | Image:SlicerIGT-VisualBlog2013.png| Some applications of the SlicerIGT extension for image-guided and navigated medical interventions. For more information see the [http://www.slicerigt.org Slicer IGT website]. | ||
| + | Image:SPV1.png|[[Documentation/Nightly/Extensions/ShapePopulationViewer|ShapePopulationViewer]] is an external tool that allows the user to dynamically interact with multiple surfaces at the same time : an interesting tool for shape visualisation and comparison. It now is available as a Slicer Extension. | ||
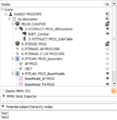
| + | Image:SlicerRT_0.13_SubjectHierarchy_ProstateEclipseLoaded.png|[http://www.na-mic.org/Wiki/index.php/2014_Winter_Project_Week:SubjectHierarchy The Subject hierarchy module and mechanism] organizes and handles loaded data, providing processing and analysis features through plugins. It aspires to be a convenient central organizing point for many of the operations that 3D Slicer and its extensions perform. | ||
| + | Image:DownloadFile 2.jpg|[[Documentation/4.3/Modules/DataStore|The Data Store]] allows a user to easily upload and download MRB files. | ||
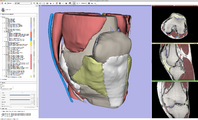
| + | Image:SlicerRT ContourRepresentations.png|[[Documentation/4.3/Modules/Contours|The Contours module]] manages contours in the RT extension package. | ||
| + | Image:MultiVolume-4.3.0-update.png| [[Documentation/4.3/Modules/MultiVolumeExplorer|MultiVolumeExplorer module]] has been updated to include the capability to plot multivolume pixel values from 2 multivolumes at the same time. This feature is particularly helpful while visualizing the results of model fits, such as those produced by the updated [[Documentation/4.3/Modules/PkModeling|PkModeling extension]]. | ||
| + | Image:MatlabBridgeLogo.png| A new extension allows to run matlab scripts without having to recompile Slicer. For more information see [[Documentation/Nightly/Extensions/MatlabBridge|'''here''']] | ||
| + | Image:ExtensionManagerScreen Shot 2013-05-23 at 9.40.23 PM.png| Extension Manager, the slicer multiplatform Appstore is open for business and free. | ||
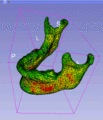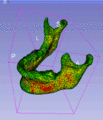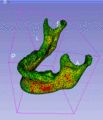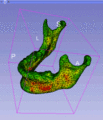
| + | Image:ScreenshotLobeSegmentation.png| Semi-automatic lobe segmentation for regional lung disease analysis as part of the Chest Imaging Platform funded by NHLBI 1R01HL116931 | ||
| + | Image:CMRToolkit.png|The [http://capulet.med.utah.edu/namic/cmrslicer Cardiac MR Extension] provides tools for segmentation and analysis of heart tissue and structure for treatment and diagnosis of Atrial Fibrillation. | ||
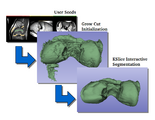
| + | Image:CarreraSliceBlogEntry.png| [[Documentation/Nightly/Modules/CarreraSliceInteractiveSegmenter|CarreraSlice]] is an interactive segmentation tool. It combines a fast implementation of the GrowCut algorithm for the 'initialization' phase with KSlice, an interactive algorithm based on Level Sets, for the 'refinement' stage. | ||
| + | </gallery> | ||
<gallery caption="2012" widths="200px" perrow="4"> | <gallery caption="2012" widths="200px" perrow="4"> | ||
| + | Image:BoxChart.png| A box plot | ||
| + | Image:2012-09-07-FetalBrain.png| Fetal brain development. The visualization is using volume rendering and the Multivolume modules in Slicer 4. Click [[media:FetalBrainGowth.mov|'''here''']] for the full movie (~24mb). Data downloaded from [http://biomedic.doc.ic.ac.uk/brain-development/index.php?n=Main.Fetal Imperial College]. | ||
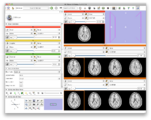
| + | Image:Screen Shot 2012-09-05 at 10.01.35 AM.png|Leveraging multi viewer layout to arrange 6 slices in the 3D viewer | ||
| + | Image:ComplexQueryWithImage.jpg|Visualization of complex query "motor system + mass + straight gyrus" using the faceted query module | ||
| + | Image:Screen Shot 2012-05-30 at 8.03.23 AM.png| Fibers colored by their mean orientation is now available in the Tractography Display module | ||
| + | Image:SlicerRT-0.2_screenshot.png|[https://www.assembla.com/spaces/slicerrt/milestones/1089853-slicerrt-0-2-release SlicerRT 0.2] has been released! [https://www.assembla.com/spaces/slicerrt/ SlicerRT] is 3D Slicer with extensions for radiotherapy research, which is developed by SparKit project with the contributions from Slicer community. | ||
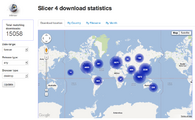
| + | Image:Screen Shot 2012-04-11 at 8.07.36 AM.png|The new extension manager | ||
| + | Image:Screen Shot 2012-04-11 at 8.11.59 AM.png|Up to date download statistics | ||
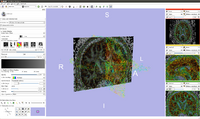
| + | Image:Screen Shot 2012-04-05 at 11.35.08 AM.png|A combination of ROI cropping of streamlines, fiducial based seeding and labelmap seeding allows to assess the deformation of streamlines which are due to tumor based deformations. | ||
| + | Image:2012-03-05-downloadpage.png|New download page which highlights the operating system of the downloading system. | ||
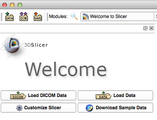
| + | Image:2012-03-04-Welcome.png|New look for the Welcome module and the "Load/Save" toolbar | ||
Image:2012-03-04-DWI2FBT.png|Step one of the DWI to Full-brain tractography wizard | Image:2012-03-04-DWI2FBT.png|Step one of the DWI to Full-brain tractography wizard | ||
Image:2012-03-04-SliceViewerControllers.png|Improved 2-stage user interface for the Slice Viewer Controllers | Image:2012-03-04-SliceViewerControllers.png|Improved 2-stage user interface for the Slice Viewer Controllers | ||
| Line 6: | Line 47: | ||
Image:2012-03-04-CompareViews.png|Updated CompareViewers and Viewer Controler Module | Image:2012-03-04-CompareViews.png|Updated CompareViewers and Viewer Controler Module | ||
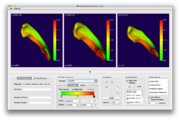
Image:Screen Shot 2012-02-23 at 7.53.51 AM.png|MultiVolumeExplorer with support for charting. See [[Documentation/Snapshot/Modules/MultiVolumeExplorer|here]] for more information. | Image:Screen Shot 2012-02-23 at 7.53.51 AM.png|MultiVolumeExplorer with support for charting. See [[Documentation/Snapshot/Modules/MultiVolumeExplorer|here]] for more information. | ||
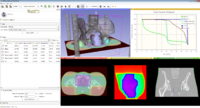
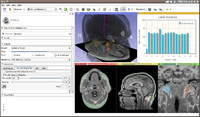
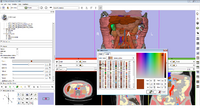
| − | Image:Screen Shot 2012-02-09 at 10.11.08 AM.png|Label Statistics with charting and visualization | + | Image:Screen Shot 2012-02-09 at 10.11.08 AM.png|[[Documentation/4.0/Modules/LabelStatistics|Label Statistics]] with charting and visualization |
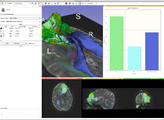
Image:3D Slicer 4.0.1.2012-02-03 180.png|Image Quantification with Label Statistics, Charting, and Volume Rendering | Image:3D Slicer 4.0.1.2012-02-03 180.png|Image Quantification with Label Statistics, Charting, and Volume Rendering | ||
Image:Slicer2011-gource.png|Frame from a movie of slicer code activity in 2011. See '''[http://boggs.bwh.harvard.edu/slicer/slicer4-2011-gource.x264.mp4 the full movie here]'''. Made with [http://code.google.com/p/gource/ gource]. Inspired by a [http://vimeo.com/33989053 an ITKv4 gource movie] | Image:Slicer2011-gource.png|Frame from a movie of slicer code activity in 2011. See '''[http://boggs.bwh.harvard.edu/slicer/slicer4-2011-gource.x264.mp4 the full movie here]'''. Made with [http://code.google.com/p/gource/ gource]. Inspired by a [http://vimeo.com/33989053 an ITKv4 gource movie] | ||
Latest revision as of 02:42, 27 November 2019
Home < Slicer4:VisualBlogThis is the Slicer 4 visual blog. Please click here if you are looking for the old Slicer 3 visual blog.
- 2016
Clinical research: 3D Slicer is used in real-time navigation of breast cancer surgery at Queen's University, Kingston, ON, Canada. The method is published in the following paper: Ungi T, Gauvin G, Lasso A, Yeo CT, Pezeshki P, Vaughan T, Carter K, Rudan J, Engel CJ, Fichtinger G. Navigated Breast Tumor Excision Using Electromagnetically Tracked Ultrasound and Surgical Instruments. IEEE Trans Biomed Eng. 2016 Mar;63(3):600-6..
- 2014
Pre-conference tutorial session Building image-guidance systems from open-source components at IEEE EMBC in Chicago (August 26, 2014).
Demo at CARS 2014 in Fukuoka, Japan. Slicer is connected to a KUKA robot for visualization of 3D models of the robot, the anatomy, and the workspace. The system is originally developed at NA-MIC Summer Project Week.
Multiplanar and volume rendered view of a CT scan of a Granny Smith apple. In plane resolution is 0.33 mm, and 1.25 mm slice thickness.
Render created in Blender of data segmented and exported from Slicer. Taimur Alavi created this from a CT scan of a friend who had broken his neck.
The TCIABrowser extension TCIABrowser gives you access to 18 collections of The Cancer Imaging Archive (http://cancerimagingarchive.net/), which contains a total of over 23,000 image series from a variety of manufacturers and of different modalities and organs.
- 2013
The Multidimensional data extension allows creating, editing, and visualizing multidimensional data sets in 3D Slicer. Browsing, replaying 4D volumes, reviewing deforming 3D models, replaying tracked ultrasound, tool navigation data in 3D, applying time-varying transforms are now all possible.
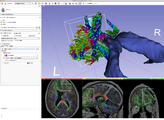
UKFTractography is a module for computing tractography of DWI images using an unscented Kalman filter. Because of its 2-tensor algorithm, it is able to model fiber crossings and capture many more fibers than a single tensor algorithm.
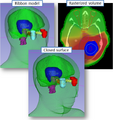
Some applications of the SlicerIGT extension for image-guided and navigated medical interventions. For more information see the Slicer IGT website.
ShapePopulationViewer is an external tool that allows the user to dynamically interact with multiple surfaces at the same time : an interesting tool for shape visualisation and comparison. It now is available as a Slicer Extension.
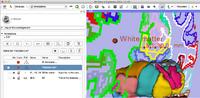
The Subject hierarchy module and mechanism organizes and handles loaded data, providing processing and analysis features through plugins. It aspires to be a convenient central organizing point for many of the operations that 3D Slicer and its extensions perform.
The Data Store allows a user to easily upload and download MRB files.
The Contours module manages contours in the RT extension package.
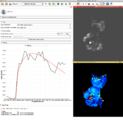
MultiVolumeExplorer module has been updated to include the capability to plot multivolume pixel values from 2 multivolumes at the same time. This feature is particularly helpful while visualizing the results of model fits, such as those produced by the updated PkModeling extension.
A new extension allows to run matlab scripts without having to recompile Slicer. For more information see here
The Cardiac MR Extension provides tools for segmentation and analysis of heart tissue and structure for treatment and diagnosis of Atrial Fibrillation.
CarreraSlice is an interactive segmentation tool. It combines a fast implementation of the GrowCut algorithm for the 'initialization' phase with KSlice, an interactive algorithm based on Level Sets, for the 'refinement' stage.
- 2012
Fetal brain development. The visualization is using volume rendering and the Multivolume modules in Slicer 4. Click here for the full movie (~24mb). Data downloaded from Imperial College.
SlicerRT 0.2 has been released! SlicerRT is 3D Slicer with extensions for radiotherapy research, which is developed by SparKit project with the contributions from Slicer community.
MultiVolumeExplorer with support for charting. See here for more information.
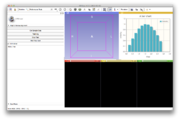
Label Statistics with charting and visualization
Frame from a movie of slicer code activity in 2011. See the full movie here. Made with gource. Inspired by a an ITKv4 gource movie
- 2011
- 2010