Difference between revisions of "Documentation/3.4"
| Line 17: | Line 17: | ||
|- | |- | ||
| rowspan="2"| | | rowspan="2"| | ||
| − | + | * The module is '''feature complete''' for the tasks advertised on 2-4-2009 | |
| − | + | * The module has a '''test'''. See [http://wiki.na-mic.org/Wiki/index.php/Slicer3:Execution_Model_Testing '''here'''] for more information. | |
| − | + | * Module has '''documentation''' on the [[Documentation-3.4#Modules|Slicer wiki]]. Please use the template provided [[Documentation-3.4#Modules|'''here''']] to structure your page. | |
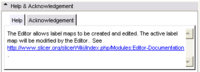
| − | + | *Please add a pointer to the documentation on the Slicer wiki to the the '''Help''' tab of the module. See the '''Editor module''' in Slicer for an example. | |
| − | + | * The contributor (and their manager/advisor), the lab (with labs/institution logo) and the funding source (with grant number, logo optional) are listed in the '''Acknowledgment''' tab of the module. Please see the '''Models module''' for and example. The people listed in the acknowledgement will be the primary people for support and maintenance relative of the module. | |
| − | + | ** '''Style Guide:''' All acknowledgment icons should be 100x100 pixels, preferably in png format. | |
| − | + | ** '''Accessing logos:''' Icons for BIRN, NAC, NA-MIC and IGT are included in Slicer3/Base/GUI//vtkSlicerBaseAcknowledgementLogoIcons.cxx/h and resources for them are in Slicer3/Base/GUI/Resources/vtkSlicerBaseAcknowledgementLogos_ImageData.h. The API for vtkSlicerModuleGUI provides access to these icons. | |
| − | + | ** '''Adding logos:''' Please add additional image resources and logo icons to these files as required in order to promote shared use (and to prevent duplication in the code.) | |
* If your module has [[Documentation-3.2|documentation in Slicer 3.2]], please copy/paste/update into the 3.4 version | * If your module has [[Documentation-3.2|documentation in Slicer 3.2]], please copy/paste/update into the 3.4 version | ||
| style="background: #e5e5e5" align="center"| Examples for the Help and | | style="background: #e5e5e5" align="center"| Examples for the Help and | ||
Revision as of 03:10, 13 February 2009
Home < Documentation < 3.4Note: This page is currently under construction
Contents
Introduction
This page is a portal for documentation about Slicer 3.4. For information for software developers, please go to the Developers page (see link in navigation box to the left).
How-To Tutorials
Feature Request and Problem Reports
We have an issues tracker for Slicer 3. You need to create an account for filing reports. We keep track of both feature requests and bug reports. Make sure to use the pull-down in the upper right to select Slicer 3.
List of Modules in need of documentation
Requirements for modules to be added to the release
|
Examples for the Help and
Acknowledgment Panels |
Main GUI
- Main Application GUI (Wendy Plesniak)
- List of Hotkeys and Keyboard Shortcuts (Wendy Plesniak)
- How to load data (Steve Pieper)
- Save Scene and Data Module (Wendy Plesniak)
Modules
- Please copy the template linked below, paste it into your page and customize it with your module's information.
Slicer3:Module_Documentation-3.4_Template
- See above for info to be put into the Help and Acknowledgment Tabs
- To put your lab's logo into a module, see here
Core and Loadable Modules
- Fetch Medical Informatics Module (Wendy Plesniak)
- Volumes Module (Alex Yarmarkovich, Steve Pieper)
- Diffusion Editor (Kerstin Kessel)
- Models Module (Alex Yarmarkovich)
- Fiducials Module (Nicole Aucoin)
- Data Module (Alex Yarmarkovich)
- Slices Module (Jim Miller)
- Transforms Module (Alex Yarmarkovich)
- Color Module (Nicole Aucoin)
- Interactive Editor (Steve Pieper)
- QDEC Module (Nicole Aucoin)
- Query Atlas Module (Wendy Plesniak)
- ROI Module (Alex Yarmarkovich)
- Volume Rendering Module (Alex Yarmarkovich)
Other Modules
Please adhere to the naming scheme for the module documentation:
- [ [Modules:MyModuleNameNoSpaces-Documentation-3.4|My Module Name With Spaces] ] (First Last Name)
Wizards
- ChangeTracker (Andriy Fedorov)
- IA FE Meshing Module (Vince Magnotta)
IGT
- OpenIGTLinkIF Module (Junichi Tokuda)
- NeuroNav Module (Haiying Liu)
- ProstateNav Module (Junichi Tokuda)
Batch processing
- EM Segementer batch (Stephen Aylward)
- Gaussian blur batch (Stephen Aylward)
- Registration batch (Stephen Aylward)
Converters
- Create a Dicom Series (Bill Lorensen)
- Dicom to NRRD (Xiaodong Tao)
- Orient Images (Bill Lorensen)
Demonstration
- Execution Model Tour (Daniel Blezek, Bill Lorensen)
- Scripted Module Example (unknown)
Filtering
- Checkerboard Filter (Bill Lorensen)
- Extract Skeleton (Pierre Seroul, Martin Styner, Guido Gerig, Stephen Aylward)
- Histogram Matching (Bill Lorensen)
- Image Label Combine (Alex Yarmarkovich)
- Resample Volume (Bill Lorensen)
- Resample Volume2 (Francois Budin)
- Threshold Image (Nicole Aucoin)
- Voting Binary Hole Filling (Bill Lorensen)
- Zero Crossing Based Edge Detection Filter
- Arithmetic
- Add Images (Bill Lorensen)
- Subtract Images (Bill Lorensen)
- Denoising
- Gradient Anisotropic Filter (Bill Lorensen checked this in)
- Curvature Anisotropic Diffusion (Bill Lorensen)
- Gaussian Blur (Julien Jomier, Stephen Aylward)
- Median Filter (Bill Lorensen)
- Morphology
- Grayscale Fill Hole (Bill Lorensen)
- Grayscale Grind Peak (Bill Lorensen)
Surface Models
- Modelmaker (Nicole Aucoin)
- Grayscale Model Maker (Bill Lorensen)
- Freesurfer Surface Section Extraction (Katharina Quintus)
- Python Surface Connectivity (Luca Antiga, Daniel Blezek)
- Python Surface ICP Registration (Luca Antiga, Daniel Blezek)
- Python Surface Toolbox (Luca Antiga, Daniel Blezek)
- Clip Model (Alex Yarmarkovich)
- Distance Transform Model (Steve Pieper)
- Model into Label Volume (Nicole Aucoin)
- Probe Volume with Model (Paint) (Lauren O'Donnell)
Python Modules
- Python Explode Volume Transform (Luca Antiga, Daniel Blezek)
- Python Script (Luca Antiga, Daniel Blezek)
- Python Numpy Script (Luca Antiga, Daniel Blezek)
Registration
- Affine Registration (Daniel Blezek)
- Deformable B-Spline Registration (Bill Lorensen)
- Diffeomorphic Demons Algorithm (Tom Vercauteren, Enger Konukoglu, Kilian Pohl)
- Linear Registration (Daniel Blezek)
- Realign Volume (Nicole Aucoin)
- Rigid Registration (Daniel Blezek)
- Test Grid Transform Registration (Yinglin Lee)
- (Utah) Deformable B-Spline Registration (Experimental) (Sam Gerber, Jim Miller, Steve Pieper, Ross Whitaker)
Segmentation
- EM Segment Command-Line (Brad Davis, Will Schroeder)
- EM Segment Simple (Brad Davis, Will Schroeder)
- EMSegmentTemplateBuilder (Brad Davis, Will Schroeder)
- Simple Region Growing (Jim Miller)
- Otsu Threshold (Bill Lorensen)
Statistics
- Calculate Volume Statistics (Tri Ngo)
- Label Statistics (Steve Pieper)
DWI and Tractography
DWI
- Estimation
- Diffusion Tensor Estimation (Raul San Jose Estepar)
- Python Diffusion Tensor Estimation (Julien von Siebenthal)
- Python Extract Baseline DWI Volume (Julien von Siebenthal)
- Filter
- Joint Rician LMMSE Image Filter (Antonio Tritan Vega, Santiago Aja Fernandez)
- Rician LMMSE Image Filter (Antonio Tritan Vega, Santiago Aja Fernandez)
- Unbiased Non Local Means filter for DWI (Antonio Tritan Vega, Santiago Aja Fernandez)
- Python Shift DWI Values (Julien von Siebenthal)
- Python Recenter Scalar to DWI Volume (Julien von Siebenthal)
- Estimation
DTI
- Resample DTI Volume (Francois Budin)
- Display (Alex Yarmakovich)
- Diffusion Tensor Scalar Measurements (Raul San Jose Estepar)
- Analysis
- Fiducial Seeding (Steve Pieper, Alex Yarmakovich)
- ROI Select (Lauren O'Donnell)
- ROI Seeding (Raul San Jose Estepar)
- Python Stochastic Tractography (Julien von Siebenthal)
Documented Modules
(please do not touch this section for now. Ron)
Main GUI
Modules
Core and Loadable Modules
CLI Modules
Modules for Downloading
We are using NITRC as a repository for contributed modules. As a general rule, we do not test them ourselves, it is the downloaders job to ensure that they do what they want them to do.
Click here to see a listing of Slicer 3 modules on NITRC.
Work is in progress to create infrastructure for searching an loading modules. See here for more information.