Difference between revisions of "Documentation/3.5"
From Slicer Wiki
| Line 178: | Line 178: | ||
*Use the Slicer Extension wizard to install extensions. Click on the icon to start the wizard | *Use the Slicer Extension wizard to install extensions. Click on the icon to start the wizard | ||
| − | *VMTK (Daniel Haehn) | + | * VMTK (Daniel Haehn) |
**You need to install the [[Modules:VMTKSlicerModule|VmtkSlicerModule]] to run any of the other three. | **You need to install the [[Modules:VMTKSlicerModule|VmtkSlicerModule]] to run any of the other three. | ||
**[[Modules:VMTKEasyLevelSetSegmentation|VMTKEasyLevelSetSegmentation]] - providing level-set segmentation of vessels, aneurysms and tubular structures using an easy interface | **[[Modules:VMTKEasyLevelSetSegmentation|VMTKEasyLevelSetSegmentation]] - providing level-set segmentation of vessels, aneurysms and tubular structures using an easy interface | ||
| Line 184: | Line 184: | ||
**[[Modules:VMTKVesselEnhancement|VMTKVesselEnhancement]] - providing vessel enhancement filters to highlight vasculature or tubular structures | **[[Modules:VMTKVesselEnhancement|VMTKVesselEnhancement]] - providing vessel enhancement filters to highlight vasculature or tubular structures | ||
| − | [[Modules:EMDTIClustering-Documentation-3.5|EM DTI Clustering]] (Mahnaz Maddah) | + | * [[Modules:EMDTIClustering-Documentation-3.5|EM DTI Clustering]] (Mahnaz Maddah) |
| + | |||
| + | * [[Modules:LabelDiameterEstimation-Documentation-3.5|Label Diameter Estimation]] (Andriy Fedorov) | ||
Revision as of 00:47, 16 November 2009
Home < Documentation < 3.5Contents
Main GUI
Main GUI
- Main Application GUI
- "Hot-keys" and Keyboard Shortcuts
- Loading Scenes and Individual Datasets through the Data Module
- Data Loading Details
- Saving Scenes and Data
- Creating and Restoring Scene Snapshots
- Extensions Management Wizard (Terry G.)
Modules
- Please copy the template linked below, paste it into your page and customize it with your module's information.
Slicer3:Module_Documentation-3.5_Template
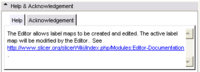
- See below for info to be put into the Help and Acknowledgment Tabs
- To put your lab's logo into a module, see here
Please adhere to the naming scheme for the module documentation:
- [ [Modules:MyModuleNameNoSpaces-Documentation-3.5|My Module Name With Spaces] ] (First Last Name)
Requirements for Modules
|
Examples for the Help and
Acknowledgment Panels |
List of Modules new to 3.5
- MRI Bias Field Correction (Nicolas Rannou, Sylvain Jaume)
- 4D Image (Viewer) (Junichi Tokuda)
- 4D Analysis (Junichi Tokuda)
- Fast Marching segmentation (Andriy Fedorov)
- Gyri Contour Segmentation (Peter Karasev)
- Subvolume extraction with ROI widget (Andriy Fedorov)
- Registration Metrics (HD and DSC) (Haytham Elhawary)
List of pre-existing Modules
Core
- Camera Module (Sebastian Barre)
- Welcome Module (Wendy Plesniak, Steve Pieper, Sonia Pujol, Ron Kikinis)
- Volumes Module (Alex Yarmarkovich, Steve Pieper)
- Diffusion Editor (Kerstin Kessel)
- Models Module (Alex Yarmarkovich)
- Fiducials Module (Nicole Aucoin)
- Data Module (Alex Yarmarkovich)
- Slices Module (Jim Miller)
- Color Module (Nicole Aucoin)
- Interactive Editor (Steve Pieper)
- ROI Module (Alex Yarmarkovich)
- Volume Rendering Module (Yanling Liu, Alex Yarmarkovich)
Specialized Modules
Please adhere to the naming scheme for the module documentation:
- [ [Modules:MyModuleNameNoSpaces-Documentation-3.5|My Module Name With Spaces] ] (First Last Name)
Wizards
- ChangeTracker (Andriy Fedorov)
- IA FE Meshing Module (Vince Magnotta)
Informatics Modules
- Fetch Medical Informatics Module (Wendy Plesniak)
- QDEC Module (Nicole Aucoin)
- Query Atlas Module (Wendy Plesniak)
Registration
- Overview:
- The Register Images module is an integrated solution to all your registration needs, if you want to have a resampled volume as output. It provides access to rigid, affine and b-spline itk technologies.
- The Transforms Module allows to manually align two volumes. This can be used for initial alignment.
- Linear, affine and Deformable B-Spline modules can be used stand-alone or one after the other. They can accept transformation matrices as the start pose and produce either transforms or resampled volumes as output.
- Transformation matrices derived from these modules can be used as input for resampling other volumes (including DTI) using the Resample Volume 2 module.
- Register Images (Stephen Aylward)
- Transforms Module (Alex Yarmarkovich)
- Linear Registration (Daniel Blezek)
- Affine Registration (Daniel Blezek)
- Deformable B-Spline Registration (Bill Lorensen)
- ACPC Transform (Nicole Aucoin)
Segmentation
- EM Segment Command-Line (Brad Davis, Will Schroeder)
- EM Segment Simple (Brad Davis, Will Schroeder)
- EM Segment Template Builder (Brad Davis, Will Schroeder)
- Simple Region Growing (Jim Miller)
- Otsu Threshold (Bill Lorensen)
Statistics
- Label Statistics (Steve Pieper)
Diffusion
DWI
- Estimation
- Diffusion Tensor Estimation (Raul San Jose Estepar)
- Python Extract Baseline DWI Volume (Julien von Siebenthal)
- Filter
- Joint Rician LMMSE Image Filter (Antonio Tristán Vega, Santiago Aja Fernandez)
- Rician LMMSE Image Filter (Antonio Tristán Vega, Santiago Aja Fernandez, Marc Niethammer)
- Unbiased Non Local Means filter for DWI (Antonio Tristán Vega, Santiago Aja Fernandez)
- Python Shift DWI Values (Julien von Siebenthal)
- Python Recenter Scalar to DWI Volume (Julien von Siebenthal)
DTI
- Resample DTI Volume (Francois Budin)
- Diffusion Tensor Scalar Measurements (Raul San Jose Estepar)
- Analysis
Tractography
- Label Seeding (Raul San Jose Estepar)
- Fiducial Seeding (Alex Yarmakovich, Steve Pieper)
- FiberBundles (Alex Yarmakovich)
- Python Stochastic Tractography (Julien von Siebenthal)
IGT
- OpenIGTLinkIF Module (Junichi Tokuda)
- NeuroNav Module (Haiying Liu)
- ProstateNav Module (Junichi Tokuda)
Filtering
- Checkerboard Filter (Bill Lorensen)
- Histogram Matching (Bill Lorensen)
- Image Label Combine (Alex Yarmarkovich)
- Resample Volume (Bill Lorensen)
- Resample Volume2 (Francois Budin)
- Threshold Image (Nicole Aucoin)
- Arithmetic
- Add Images (Bill Lorensen)
- Subtract Images (Bill Lorensen)
- Denoising
- Gradient Anisotropic Filter (Bill Lorensen checked this in)
- Curvature Anisotropic Diffusion (Bill Lorensen)
- Gaussian Blur (Julien Jomier, Stephen Aylward)
- Median Filter (Bill Lorensen)
- Morphology
- Voting Binary Hole Filling (Bill Lorensen)
- Grayscale Fill Hole (Bill Lorensen)
- Grayscale Grind Peak (Bill Lorensen)
Surface Models
- Modelmaker (Nicole Aucoin)
- Grayscale Model Maker (Bill Lorensen)
- Freesurfer Surface Section Extraction (Katharina Quintus)
- Python Surface Connectivity (Luca Antiga, Daniel Blezek)
- Python Surface ICP Registration (Luca Antiga, Daniel Blezek)
- Python Surface Toolbox (Luca Antiga, Daniel Blezek)
- Clip Model (Alex Yarmarkovich)
- Model into Label Volume (Nicole Aucoin)
Batch processing
- EM Segmenter batch (Julien Jomier, Brad Davis)
- Gaussian Blur batch (Julien Jomier, Stephen Aylward)
- Register Images batch (Julien Finet, Stephen Aylward)
- Resample Volume batch (Julien Finet)
Converters
- Create a Dicom Series (Bill Lorensen)
- Dicom to NRRD (Xiaodong Tao)
- Orient Images (Bill Lorensen)
- Python Explode Volume Transform (Luca Antiga, Daniel Blezek)
- Extract Subvolume (Steve Pieper)
Slicer Extensions
- Slicer Extensions are a mechanism for third parties to provide modules which extend the functionality of 3d Slicer.
- Some of the extensions do not use the Slicer license. Please review carefully.
- For a subset of extensions, you can use the extension wizard in Slicer to find their webpages and to install/uninstall individual extensions. In case of problems with those modules, please talk directly to the developers of the extensions.
- The version that is available through the extension manager is chosen by the developer of that extension
- Extensions are compiled as part of the nightly build. In order to have your extension compiled nightly and made available to end users, please contact the Slicer team.
Installation
- Use the Slicer Extension wizard to install extensions. Click on the icon to start the wizard
- VMTK (Daniel Haehn)
- You need to install the VmtkSlicerModule to run any of the other three.
- VMTKEasyLevelSetSegmentation - providing level-set segmentation of vessels, aneurysms and tubular structures using an easy interface
- VMTKLevelSetSegmentation - providing level-set segmentation of vessels, aneurysms and tubular structures using different algorithms
- VMTKVesselEnhancement - providing vessel enhancement filters to highlight vasculature or tubular structures
- EM DTI Clustering (Mahnaz Maddah)
- Label Diameter Estimation (Andriy Fedorov)