Documentation/Nightly/Announcements
|
For the latest Slicer documentation, visit the read-the-docs. |
| Summary | What is 3D Slicer | Slicer Nightly Highlights | Slicer Training | Slicer Extensions | Other Improvements, Additions & Documentation |
Summary
The community of Slicer developers is proud to announce the release of Slicer Nightly.
The development of Slicer, including its numerous modules, extensions, datasets, patches sent on user and developer lists, issues reports, suggestions, ... is made possible by awesome users, developers, contributors, commercial partners from around the world and also invaluable grants and funding agencies.
For more details, see Acknowledgments page.
- Slicer Nightly introduces
- Close to 700 feature improvements and bug fixes have resulted in improved performance and stability
- Dozens of new and improved core modules and extensions
- Click here to download Slicer Nightly for different platforms and find pointers to the source code, mailing lists and the bug tracker.
- Please note that Slicer continues to be a research package and is not intended for clinical use. Testing of functionality is an ongoing activity with high priority, however, some features of Slicer are not fully tested.
- The Slicer Training page provides a series of tutorials and data sets for training in the use of Slicer.
- slicer.org is the portal to the application, training materials, and the development community.
What is 3D Slicer
3D Slicer is:
- A software platform for the analysis (including registration and interactive segmentation) and visualization (including volume rendering) of medical images and for research in image guided therapy.
- A free, open source software available on multiple operating systems: Linux, MacOSX and Windows
- Extensible, with powerful plug-in capabilities for adding algorithms and applications.
Features include:
- Multi organ: from head to toe.
- Support for multi-modality imaging including, MRI, CT, US, nuclear medicine, and microscopy.
- Bidirectional interface for devices.
There is no restriction on use, but Slicer is not approved for clinical use and intended for research. Permissions and compliance with applicable rules are the responsibility of the user. For details on the license see here
Citing Slicer
To acknowledge 3D Slicer as a platform, please cite the Slicer web site (http://www.slicer.org) and the following publication:
Fedorov A., Beichel R., Kalpathy-Cramer J., Finet J., Fillion-Robin J-C., Pujol S., Bauer C., Jennings D., Fennessy F., Sonka M., Buatti J., Aylward S.R., Miller J.V., Pieper S., Kikinis R. 3D Slicer as an Image Computing Platform for the Quantitative Imaging Network. Magnetic Resonance Imaging. 2012 Nov;30(9):1323-41. PMID: 22770690.
Slicer Nightly Highlights
- New and Improved Modules
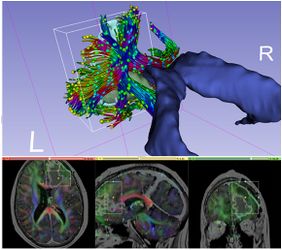
- Introduced SlicerDMRI extension including diffusion MRI modules formerly in Slicer core and new functionality:
- DICOM tractography import/export
- Improved diffusion brain mask generation
- UKF multi-fiber tractography now available on Windows
- Improved user interface and documentation
- Segments.jpg
New Segments Module and Segment Editor
- ScreenCapture.jpg
Make movies and animated gifs directly from the Slicer UI
- OpenGL2.jpg
New VTK versions including new OpenGL2 backend for more modern and efficient rendering
- Tables.jpg
Spreadsheet like tables for quantitative results
- DICOM.jpg
Improved support for DICOM objects using the latest DCMTK
Slicer Training
The Slicer Training page provides a series of updated tutorials and data sets for training in the use of Slicer Nightly.
The first hands-on training and showcase events using Slicer 4.6 will be organized at the Memorial Sloan Kettering Cancer Center, New York City, NY and the annual meeting of the Radiological Society of North America (RSNA 2016), Chicago, Il.
- New Tutorials
The Subject Hierarchy tutorial demonstrates the basic usage and potential of Slicer’s data manager module Subject Hierarchy using a two-timepoint radiotherapy phantom dataset. NEW
The Fiber Bundle Selection and Scalar Measurements tutorial guides through the use of the Diffusion Bundle Selection module and the Fiber Tract Scalar Measurement module for diffusion MRI tractography data analysis. NEW
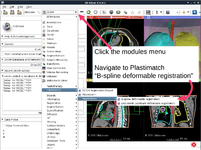
The Plastimatch tutorial guides through registration and wrapping of DICOM and DICOM-RT data using the Plastimatch extension of 3D Slicer. NEW
3D Slicer YouTube channel has been reorganized. A new video 'Zooming into microscopy data using 3D Slicer' developed by the community has been added to the channel NEW
Slicer Extensions
- New Extensions
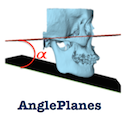
AnglePlanes This Module is used to calculate the angle between two planes by using the normals NEW
SlicerOpenCV This Module provides a way for a Slicer extension developer use the OpenCV pacakge. NEW
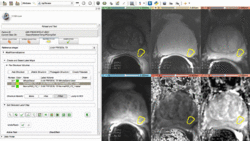
New mpReview extension to support review and annotation of multiparametric image data. The driving use case for the development of this module was review and segmentation of the regions of interest in prostate cancer multiparametric MRI.
New SliceTracker extension to support navigation and guidance during in-bore MRI-guided prostate biopsy. Main documentation for this module is hosted on Gitbook - give us your feedback about this approach for documenting Slicer functionality! https://fedorov.gitbooks.io/slicetracker/content/
New SlicerPathology extension for tools for automatic and semi-automatic pathology image segmentation, with the interface to caMicroscope.
Improved Extensions in Slicer Nightly
- Improved SlicerProstate extension.
- improved reporting of DWI model fit diagnostics
- refactoring of the code to separate functionality common to mpReview and SliceTracker extensions into a reusable library
Extensions removed from Slicer Nightly
Extensions renamed
Other Improvements, Additions & Documentation
Optimization
Transforms
DICOM
Data processing
CLI
Usability
SubjectHierarchy
Python scripting
Editor
Markups
LabelMapVolumeNode
Slice viewers
DataProbe
SliceViewAnnotations
OpenIGTLink
For Developers
Modules and Extensions
Slicer Core
- Improved Toolkits
Moved from OpenIGTLink 66e272d to 849b434 (53 commits)
Looking at the Code Changes
From a git checkout you can easily see the all the commits since the time of the 4.5.0-1 release:
git log v4.5.0-1..HEAD
To see a summary of your own commits, you could use something like:
git log v4.5.0-1..HEAD --oneline --author=me
see the git log man page for more options.
Commit stats and full changelog