Documentation/Nightly/Modules/DiffusionWeightedVolumeMasking
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on NA-MIC can be obtained from the NA-MIC website. | |||||||||||
|
Module Description
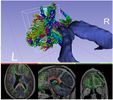
Creates a brain mask from a diffusion weighted image volume. The mask can be used during diffusion tensor estimation or tractography seeding.
The brain mask is computed by averaging all baseline (non-diffusion-weighted) images, applying the Otsu thresholding algorithm to segment tissue voxels, and then removing small unconnected regions.
Use Cases
Most frequently used for these scenarios:
- Use Case 1:
- Use Case 2:
Tutorials
Links to tutorials that use this module
Panels and their use
Parameters:
- IO: Input/output parameters
- Input DWI Volume (inputVolume): Input DWI volume
- Output Baseline Volume (outputBaseline): Extracted baseline volume
- Output Diffusion Brain Mask (thresholdMask): Output Diffusion Brain Mask
- Mask Settings:
- Baseline B-Value Threshold Parameter (baselineBValueThreshold): Volumes with B-value below this threshold will be considered baseline images and included in mask calculation.
- Remove Islands in Brain Mask (removeIslands): Removes disconnected regions from brain mask.
List of parameters generated transforming this XML file using this XSL file. To update the URL of the XML file, edit this page.
Similar Modules
- Point to other modules that have similar functionality
References
Publications related to this module go here. Links to pdfs would be useful. For extensions: link to the source code repository and additional documentation
Information for Developers
| Section under construction. |