Documentation/4.5/Modules/DiceComputation
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work is supported by NA-MIC, NCIGT, and the Slicer Community. | |||||||
This project is supported by National Institute of Health (5P01CA067165, 5R01CA124377, 5R01CA138586, 2R44DE019322, 7R01CA124377, 5R42CA137886, 8P41EB015898). |
Module Description
DiceComputation is capable of calculating the Dice Similarity Coefficient (DSC) between multiple label map images. If the label map images represent registered segmented structures, the DSC will provide a volumetric alignment between the images. The DSC gives measures of the volumetric overlap between two of the segmented structures, and indicates twice the number of voxels which are shared by or are common to both structures divided by the total number of non-zero voxels in both structures. The DSC can range from zero to one, where zero is no alignment between images and one is perfect alignment.
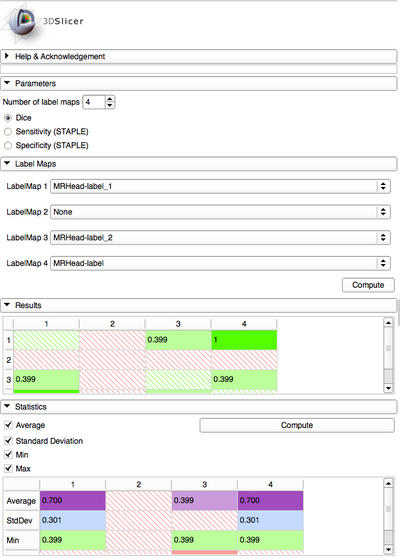
If more than 2 label map images are specified as input, the module computes DSCs for all combinations and present results in a NxN matrix, where N is the number of the label map images, with color coding for convenient reading.
Use Cases
DiceComputation compute DSC to quantitatively compare several registered segmented volumes. It could be used to compare:
- Several algorithms with ground truth
- Automated segmentation vs manual
- etc...
Tutorials
N/A
Panels and their use
- Parameters
- Number of label map images to use for computation
- Label Maps
- Select labels map images (only label maps could be selected) to use for computation or None
- Compute Dice coefficient: Press button to start computation
- Results
- Once computation is over, results will appear in an NxN matrix with color coding
- Red stripes indicate no label map have been selected (None)
- Green stripes indicate this is Dice's coefficient of a label map with itself (always 1.0, so not displayed)
- Values represent Dice's similarity coefficient. The bigger the value is, the greener the background is, in order to quickly find higher matches.
- Once computation is over, results will appear in an NxN matrix with color coding
Similar Modules
N/A
References
- A. Bharatha, M. Hirose, N. Hata, S. K. Warfield, M. Ferrant, K. H. Zou, E. Suarez-Santana, J. Ruiz-Alzola, A. D'Amico, R. A. Cormack, R. Kikinis, F. A. Jolesz, and C. M. Tempany, "Evaluation of three-dimensional finite element-based deformable registration of pre- and intraoperative prostate imaging," Med Phys, vol. 28(12), pp. 2551-60, 2001.
Information for Developers
| Section under construction. |