Documentation/Nightly/Modules/SobolevSegmenter
Introduction and Acknowledgements
|
This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research. Information on NA-MIC can be obtained from the NA-MIC website. | |||
|
Module Description
This extension implements Sobolev inner product based active contour, using Chan-Vese energy functional. The segmentation is appropriate for 2D images. The obtained parametric contour is generally smooth, but able to catch concavities.
Use Cases
The Sobolev segmenter is a general image segmenter, and it can be used with any 2D data, as explained in the tutorial.
Tutorials
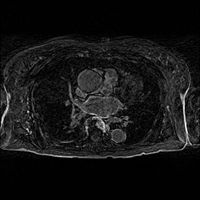
- Load the image (input volume):
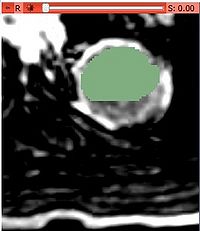
- Use built in editor to select a single initial mask (or load a binary mask file):
- Select Segmenation->SobolevSegmenter module
- Choose the Input Volume and the Initial Mask accordingly. Create a new volume for the Output Volume.
- Press Apply button.
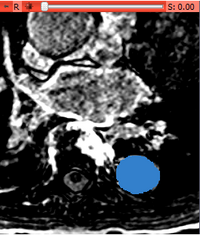
- After a few second the following output volume should appear:
Panels and their use
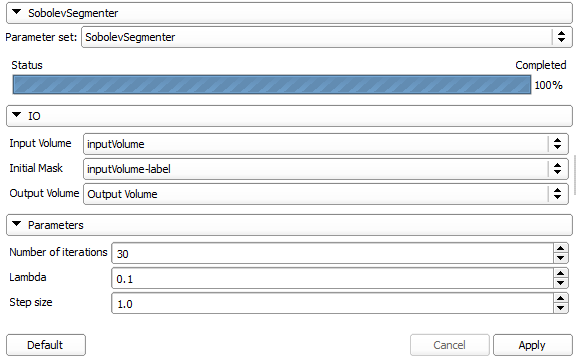
The module has the following panel:
The IO section of this panel defines two input images (data and initial mask) and one output image (final mask). The algorithm has three parameters: self-explanatory number of iterations and contour evolution step size. In addition, the parameter lambda chooses the smoothness of the contour (smoothing kernel width).
Similar Modules
N/A
References
N/A
Information for Developers
| Section under construction. |