EMSegmenter-Tasks:Template
Return to EMSegmenter Task Overview Page
Contents
This wiki template can be used for new tasks
Description
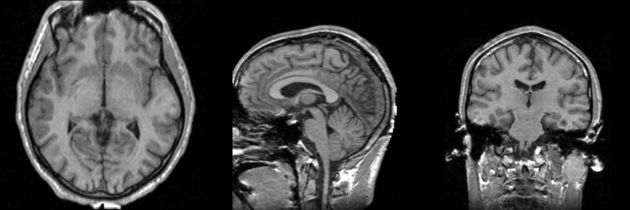
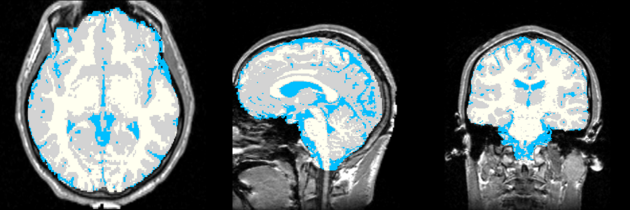
Single channel automatic segmentation of t1w-MRI brain scans into the major tissue classes (gray matter, white matter, csf). The task can only be applied to t1w brain scan showing parts of the skull and neck. The pipeline consist of the following steps:
- Step 1: Perform image inhomogeneity correction of the MRI scan via N4ITKBiasFieldCorrection (Tustison et al 2010)
Anatomical Tree
- root
- background (BG)
- air (AIR)
- intracranial cavity (ICC)
- white matter (WM)
- background (BG)
Atlas
- how did you construct the atlas ?
-- what did you do for skull stripping or any other preprocessing
-- What did you do for group registration
-- how did you generate the segmentations
-- how many set of images did you use
-- where did you get the data from
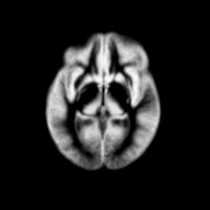
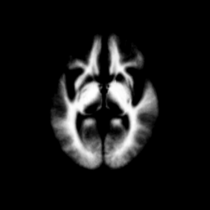
Atlas was generated based on 82 scans and corresponding segmentations provided by Psychiatry Neuroimaging Laboratory, BWH. We registered the scans to a preselected template via Warfield et al. 2001.
Image Dimension = 256 x 256 x 124
Image Spacing = 0.9375 x 0.9375 x 1.5
|
|
|
|
| Template (T1) | CSF | GM | WM |
Result
Acknowledgment
The construction of the pipeline was supported by funding from NIH NCRR 2P41RR013218 Supplement.
Citations
- Pohl K, Bouix S, Nakamura M, Rohlfing T, McCarley R, Kikinis R, Grimson W, Shenton M, Wells W. A Hierarchical Algorithm for MR Brain Image Parcellation. IEEE Transactions on Medical Imaging. 2007 Sept;26(9):1201-1212.