Stanford Simbios group
Contents
Aim
To develop a generic semi-automatic segmentation toolkit to convert images of musculoskeletal structures to 3D models.
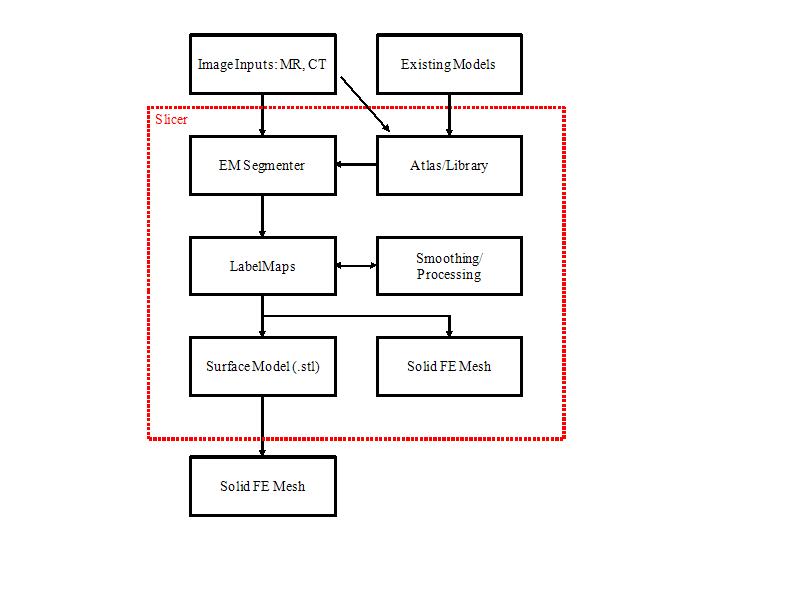
Process Flowchart
Progress
Atlas Generation from Input MR Images
Model Generation from Input MR Images
Pre-Segmented models for femur, patella and tibia are obtained for a patient (in .stl format). This models can be prepared using the hypermesh software. The models contain vertices of triangles representing femur, tibia and patella regions.
Create a filled label map using PolyDataToFilledLabelMap module in slicer
The models generated in the above step are closed but hollow. But EM Segmentation requires the atlas given to be in closed filled format. Hence we converted the hollow closed models into filled label volumes using the module PolyDataToFilledLabelMap module in Slicer. The output label map of this module is of datatype unsigned char and is converted to short datatype.
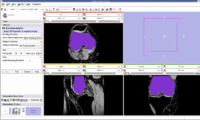
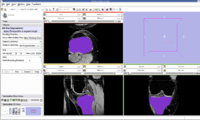
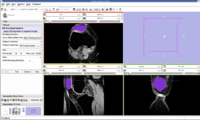
EM Segmentation based on the atlas
The output label map in the above is given as input atlas in the EM Segmentation step. As an initial step we ran EM Segmentation on the same patient. The EM Segmented output is given in the below figure.
Register Images
Register Images Module in Slicer
We are now in the process of trying to register new patient image to the existing patient image. If we register the images, we can get the transform which can be used to register the existing atlas (label map) to the new patient.
Multi Image Registration
We have also tried the approach of Non-rigid Groupwise Registration using Bspline Deformation Model by Serdar K. Balci, Polina Golland and William M. Wells. Here are the results of their approach.
Datasets
Below are the datasets that we used in our experiments. All the images are in standard dicom format.