Documentation/Nightly/Extensions/3D Model Segmentation
From Slicer Wiki
Home < Documentation < Nightly < Extensions < 3D Model Segmentation
It is our intention to add semi-automatic and automatic segmentation tools based on deep learning and other machine learning methods to this module in the future. Check back in sometime soon!
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Module Description
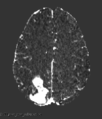
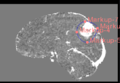
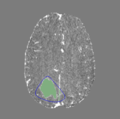
The 3D Model Segmentation module allows users to quickly create smooth, 3D regions of interest with only a few clicks, and then provides a variety of tools to threshold and refine those regions of interest. The module is broken up into several steps, which the user can progress through with "Back" and "Next" buttons on the bottom of the module panel. The module can be used on single volumes, or on pre- and post-contrast pairs. In the latter case, utilities for registering, normalizing, and subtracting volumes are provided. Check the tutorials section for some usage examples, and send any bugs, comments, or feature requests to abeers@mgh.harvard.edu.It is our intention to add semi-automatic and automatic segmentation tools based on deep learning and other machine learning methods to this module in the future. Check back in sometime soon!
Use Cases
- Tissue segmentation with and pre- and post-contrast images.
- Intensity thresholding for parametric maps.
- Subtraction map creation and registration.
- Adaptive volume cropping
Tutorials
https://www.youtube.com/watch?v=_7oZygGp2ds&t=13s
Documentation
Step 1. Volume Selection
Step 2. Registration (Optional)
Step 3. Normalization and Subtraction (Optional)
Step 4. ROI
Step 5. Thresholding
Step 6. Review
Similar ModulesVolume Clip - https://www.slicer.org/wiki/Documentation/Nightly/Extensions/VolumeClip ReferencesNone Information for Developers
|