Documentation:Nightly:Registration:RegistrationLibrary:RegLib C38
From Slicer Wiki
Home < Documentation:Nightly:Registration:RegistrationLibrary:RegLib C38
Contents
Slicer Registration Library Case 38: TBI
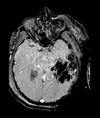
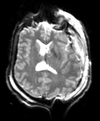
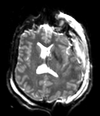
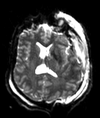
Input
|
|
|
|

|
|
|
|
|
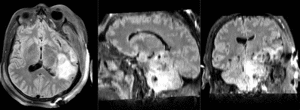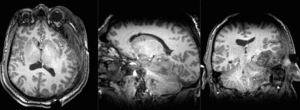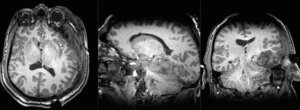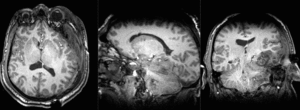
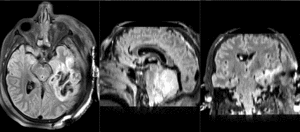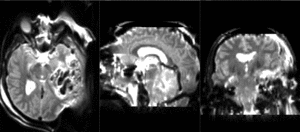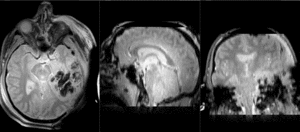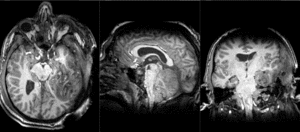
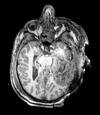
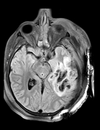
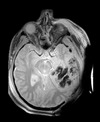
| T1pre | FLAIR | T1post | coronal T2 | SWI | EP50 | EP75 | EP100 |
Modules used
Download (from NAMIC MIDAS)
- RegLib_C38.mrb (input data, Slicer mrb file. 42 MB).
Video Screencasts
Key Strategies
- we register all the scans within each exam to the T1
- we limit registration degree of freedom to 6 DOF (rigid transformation only).
Procedure
- Load all datasets via drag&drop or via "AddData / AddVolume" ... do not center the volumes, since they have reasonable alignment at the outset
- the registration procedure below is identical for all other scans in the sequence, i.e. just replace the Moving Volume and the Slicer Linear Transform", everything else can stay at default
- open the General Registration (BRAINS) module
- Fixed Image Volume: T1
- Moving Image Volume: T2
- Output Settings:
- Slicer BSpline Transform": none
- Slicer Linear Transform: create new transform, rename to "Xf1_T2"
- Output Image Volume: none
- Registration Phases: check box for Rigid only
- leave all other settings at defaults
- click: Apply; runtime < 10 sec (MacPro QuadCore 2.4GHz)
- this will generate the alignment transform. Note that the Slicer4.1 version of BRAINS registration does not automatically place the moving volume inside the result transform, as it did for Slicer 3.6. In order to see the result, you must either go to the Data module and drag the moving volume inside the transform node, or when calculating the registration, select to generate a resampled Output Volume. The demo screencast above shows how to apply the result transforms without resampling and then saving the images under a new name. It also shows where in the saved image the new orientation is stored.