Announcements:Slicer3.4
Contents
Introduction
The community of Slicer developers is proud to announce the release of Slicer 3.4.
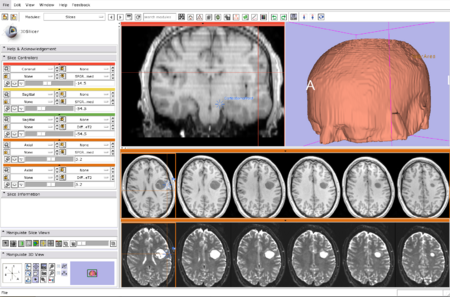
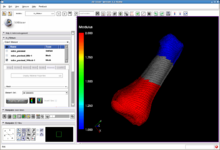
Slicer 3.4 is a general purpose biomedical computing application with extensive built-in visualization and analysis capabilities, accessible through an easy to use graphical interface.
New Features in Slicer 3.4
Slicer 3.4 has many new features! Please also note that many of these new features are still being developed and will be improved in future releases, so you may encounter some unexpected behavior. If you do encounter unexpected behavior, you can join the Slicer users' mailing by sending email to slicer-users-request@massmail.spl.harvard.edu with the subject "subscribe", and report the behavior there to see if there is a work around (or check the archives), and to the bug tracker if it needs to be solved by the developers.
Click here to download different versions of Slicer3 and find pointers to the source code, mailing lists and bug tracker. Please note that Slicer continues to be a research package and is not intended for clinical use. Testing of functionality is an ongoing activity with high priority, however, some features of Slicer3 are not fully tested.
New Features
- Welcome Module at Startup
- Improved Save Module
- Improved Editor
- New Registration module
- New Slices module
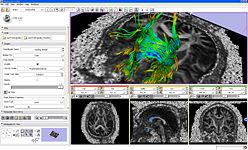
- Fiducial based tractography
- Improved SceneSnapshot Screen Capture functionality
- Compare View and Cross Hairs
- Support for Extension Server for installing plug-ins
- Improved Dicom Support
- MRML scenes and all data load from and save into XNAT desktop
Links to Latest Slicer 3.4 Binaries
- 64-bit Linux
- 32-bit Linux
- Darwin PPC
- Darwin x86
- Windows
Highlights
- Slicer v3.4 - New and Improved Feature Highlights
Reporting Bugs
Please go to http://www.na-mic.org/Bug/ and report bugs for Slicer version 3.
Where to go for Help
Compilation/development questions can be sent to the Slicer Developers Mailing List. Questions about running Slicer can be sent to the Slicer Users Mailing List.
Subscription management tools and archives are available at:
http://massmail.bwh.harvard.edu/mailman/listinfo/slicer-devel
http://massmail.bwh.harvard.edu/mailman/listinfo/slicer-users
Acknowledgments
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on the National Centers for Biomedical Computing can be obtained from http://nihroadmap.nih.gov/bioinformatics. This work was also supported by NIH grant P41 RR13218 (NAC) and the Biomedical Informatics Research Network.