Modules:ResampleScalarVectorDWIVolume-Documentation-3.6
Return to Slicer 3.6 Documentation
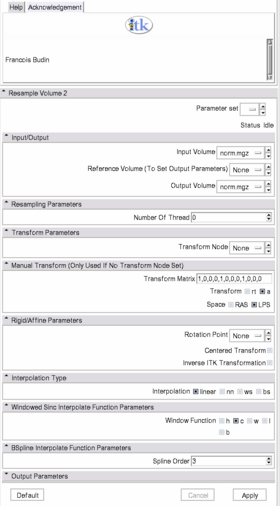
Resample Scalar/Vector/DWI Volume
 Original Image [1] |
General Information
Module Type & Category
Type: Interactive or CLI
Category: Base or (Filtering, Registration, etc.)
Authors, Collaborators & Contact
- Author1: Affiliation & logo, if desired
- Contributor1: Affiliation & logo, if desired
- Contributor2: Affiliation & logo, if desired
- Contact: name, email
Module Description
Overview of what the module does goes here.
Usage
Examples, Use Cases & Tutorials
- Note use cases for which this module is especially appropriate, and/or link to examples.
- Link to examples of the module's use
- Link to any existing tutorials
Quick Tour of Features and Use
List all the panels in your interface, their features, what they mean, and how to use them. For instance:
- Input panel:
- Parameters panel:
- Output panel:
- Viewing panel:
Development
Dependencies
The module uses some source code present in the ResampleDTI module repository.
Known bugs
Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Customize following links for your module:
Source code:
Source code from ResampleDTI
- itkWarpTransform3D.h
- itkWarpTransform3D.txx
- itkTransformDeformationFieldFilter.h
- itkTransformDeformationFieldFilter.txx
- deformationfieldio.h
- deformationfieldio.cxx
- dtitypes.h
- itkHFieldToDeformationFieldImageFilter.h
- itkHFieldToDeformationFieldImageFilter.txx
Doxygen documentation:
More Information
Acknowledgment
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on the National Centers for Biomedical Computing can be obtained from http://nihroadmap.nih.gov/bioinformatics.
References
Publications related to this module go here. Links to pdfs would be useful.