Difference between revisions of "Modules:GaussianBlurBatch-Documentation-3.4"
| Line 4: | Line 4: | ||
Gaussian Blur BatchMake | Gaussian Blur BatchMake | ||
{| | {| | ||
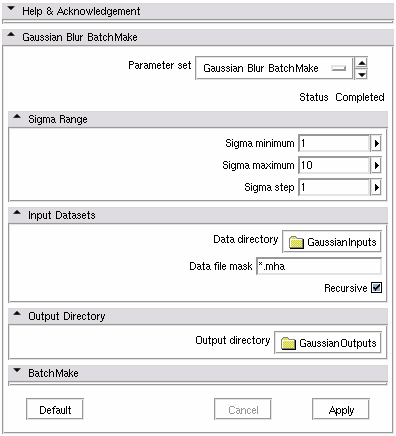
| − | |[[Image: | + | |[[Image:GaussianBatchMakeModule.png|thumb|580px|Gaussian Blur BatchMake GUI]] |
|} | |} | ||
Revision as of 19:23, 6 April 2009
Home < Modules:GaussianBlurBatch-Documentation-3.4Return to Slicer 3.4 Documentation
Module Name
Gaussian Blur BatchMake
General Information
Module Type & Category
Type: CLI
Category: Filtering
Authors, Collaborators & Contact
- Author: Stephen Aylward
- Contact: stephen.aylward at kitware.com
Module Description
Convolves the image with a Gaussian kernel wherein the Gaussian has a standard deviation specified by the user (GUI field "sigma") and the kernel width in each dimension is 6 times the standard deviation of the Gaussian.-
Usage
Input volume = <grayscale image> Output volume = <new image> Sigma = <std dev of gaussian for convolution>
Examples, Use Cases & Tutorials
- Use to reduce noise
Quick Tour of Features and Use
- Set the sigma value to control the amount of blur. Larger sigma = more blur.
Development
Dependencies
None
Known bugs
Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Source Code: GaussianBlurBatchMakeModule.cxx
XML Description: GaussianBlurBatchMakeModule.xml
Documentation:
More Information
Acknowledgment
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on the National Centers for Biomedical Computing can be obtained from National Centers for Biomedical Computing.