Difference between revisions of "Documentation/Nightly/Training"
Tag: 2017 source edit |
|||
| Line 30: | Line 30: | ||
{| width="100%" | {| width="100%" | ||
| | | | ||
| − | *The [https://www.dropbox.com/s/vn8sqlof2kag2kk/SlicerWelcome-tutorial_Slicer4.8_SoniaPujol.pdf?dl=0 | + | *The [https://www.dropbox.com/s/vn8sqlof2kag2kk/SlicerWelcome-tutorial_Slicer4.8_SoniaPujol.pdf?dl=0 Slicer Welcome tutorial] is an introduction to Slicer. |
*Author: Sonia Pujol, Ph.D. | *Author: Sonia Pujol, Ph.D. | ||
*Audience: First-time users who want a general introduction to the software | *Audience: First-time users who want a general introduction to the software | ||
| Line 359: | Line 359: | ||
| style="width:20%" |[[Image:3DPrinting.png|right|250px|]] [https://www.youtube.com/watch?v=MKLWzD0PiIc Preparing data for 3D printing - Author: Nabgha Farhat] | | style="width:20%" |[[Image:3DPrinting.png|right|250px|]] [https://www.youtube.com/watch?v=MKLWzD0PiIc Preparing data for 3D printing - Author: Nabgha Farhat] | ||
| style="width:20%" |[[Image:DICOM2.png|right|250px|]] [https://www.youtube.com/watch?v=nzWf4xHy1BM& How to export CT and segmentation data to DICOM - Author: Andras Lasso, Csaba Pinter] | | style="width:20%" |[[Image:DICOM2.png|right|250px|]] [https://www.youtube.com/watch?v=nzWf4xHy1BM& How to export CT and segmentation data to DICOM - Author: Andras Lasso, Csaba Pinter] | ||
| − | | style="width:20%" |[[Image:LocalThresholdEffect.png|right|250px|]] [https://www.youtube.com/watch?time_continue=26&v=cevlMLyhfK8&feature=emb_logo | + | | style="width:20%" |[[Image:LocalThresholdEffect.png|right|250px|]] [https://www.youtube.com/watch?time_continue=26&v=cevlMLyhfK8&feature=emb_logo Local Threshold Effect - Author: Kyle Sunderland] |
|- | |- | ||
|} | |} | ||
| Line 406: | Line 406: | ||
*[https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247486025&idx=1&sn=b281324893be4ab116d20826f1b426c3&chksm=eacc007bddbb896d9deb096f209278f40c0b52c6410a8a9ff3ce8c3697c99304f18eb678f11e&mpshare=1&scene=24&srcid=02125v1kxvIGmfkxx7mUZcCM#rd Mobile phone positioning and AR application 手机定位及AR应用的初步探索] | *[https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247486025&idx=1&sn=b281324893be4ab116d20826f1b426c3&chksm=eacc007bddbb896d9deb096f209278f40c0b52c6410a8a9ff3ce8c3697c99304f18eb678f11e&mpshare=1&scene=24&srcid=02125v1kxvIGmfkxx7mUZcCM#rd Mobile phone positioning and AR application 手机定位及AR应用的初步探索] | ||
| + | *Brain tumor localization [https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247483699&idx=1&sn=5bdb822dc7b93d4b0ebde2e8abdebe2d&chksm=eacc0b01ddbb821704929fa61c3d2995ad1aa3e9dfad2ee4d53fcd681ebf4bde6bb43becef08#rd 3DSlicer实战之浅表肿瘤定位 霍贵通] | ||
| + | *Localization of intracranial lesions [https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247483717&idx=1&sn=8ca6a245abdb54617c021e50573dd865&chksm=eacc0b77ddbb82610d5838236331f52b7ec96c587898d972236e9cf08b6f3c228c9c5e16e9fe#rd 颅内病变体表定位 束旭俊] | ||
| + | *CPA tumor: surgical approach simulation [https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247483805&idx=1&sn=fd315dfc75dc2ad6f2fc281f7871586b&chksm=eacc0bafddbb82b9bff78fc63c9d80af1c27e70cac298647ca63541d3596d5ea1936e92a969a#rd 3Dslicer模拟手术入路之CPA肿瘤] | ||
| + | *Neuroendoscopic surgery for cerebral hemorrhage [https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247483863&idx=1&sn=ecd46c711c30435ff1fba6e63af3f8d0&chksm=eacc0be5ddbb82f327c99e47d3af887b4c63d66b2f2d776d6965cde0033bfef06881af5e5284#rd 3D Slicer在脑出血神经内镜手术中的应用 谢国强] | ||
| + | *Parasinus meningiomas [https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247483870&idx=1&sn=2876f32e99da710fe84e588ba6cd5219&chksm=eacc0becddbb82faa952ab565562c13258619cfbb6505b55135969512291a9db912ead52672d#rd 3D Slicer临床应用之窦旁脑膜瘤 霍显浩] | ||
| + | *Using 3D Slicer to merge images from different examination sites [https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247483912&idx=1&sn=a87d27e861cfc601bdaffea3714cdd99&chksm=eacc083addbb812c5b7fb37398e8e253534f7d86ab77f55da2085045a1b81b1bcbcd99708bed#rd 利用3D Slicer融合不同检查部位的图像 王奎重] | ||
| + | *KiwiViewer—[https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247483948&idx=1&sn=05b4683ab126513554a593a4a29f415d&chksm=eacc081eddbb81080bb1ba6c7c69b13f1d4d2b140f8df19c7860164ef9e16d7698ad1a434139#rd 将患者的医学影像装进口袋 束旭俊] | ||
| + | *Data matching [https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247484003&idx=1&sn=baf76f3bdb37c8ceb6faba34b018d87a&chksm=eacc0851ddbb8147cd174da3c3d05ca375398758547f36357f8717edb4318e91eeea1b0c51b3#rd 3D Slicer教程之数据匹配(一) 霍贵通] | ||
| + | *Application of 3DSlicer Virtual Endoscope in Cerebral Hemorrhage [https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247484034&idx=1&sn=81c5e566cc58ede4a450c8c6c346d1b6&chksm=eacc08b0ddbb81a6d148327eb32a69524df501276a55e2aa5131766022782f708cc376e8978f#rd 3Dslicer虚拟内镜在脑出血清除术演练中的应用 李珍珠] | ||
| + | *GyroGuide [https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247484051&idx=1&sn=56ecbb57d2e14c8f95c2663cce6012fc&chksm=eacc08a1ddbb81b7f3f65f0cf01ae8f6ab419c7d94e0fadef13ef7ac1ba4013a1ff63c0764fb#rd 的使用方法 束旭俊] | ||
| + | *Basics of Multimodal Fusion/Data Matching in 3DSlicer [https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247484085&idx=1&sn=ed68e6fe2218974f515f148eaa0e6fe1&chksm=eacc0887ddbb819199bbc747509493b3f9f34c33e32ca25a2e72ab2b3861a9f07a590eec7997#rd 多模态融合基础篇——3DSlicer教程之数据匹配(二) 霍贵通] | ||
| + | *Intracranial hematoma aspiration [https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247484105&idx=1&sn=7a82e3a46103a4a879b9c6ba9210485e&chksm=eacc08fbddbb81ed5a12c74dd039caf08df84508c77dadddbb6d92a259aa227750de6e01fb5a#rd 3Dslicer实战之简易导航下颅内血肿碎吸术 赵端允] | ||
| align="right" | | | align="right" | | ||
Revision as of 22:26, 4 March 2020
Home < Documentation < Nightly < Training
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction: Slicer Nightly Tutorials
- This page contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment (version Nightly release) to accomplish certain tasks.
- For tutorials for other versions of Slicer, please visit the Slicer training portal.
- For "reference manual" style documentation, please visit the Slicer Nightly documentation page
- For questions related to the Slicer4 Training Compendium, please send an e-mail to Sonia Pujol, Ph.D., Director of Training of 3D Slicer.
- Some of these tutorials are based on older releases of 3D Slicer and are being upgraded to Slicer4.10. The concepts are still useful but some interface elements and features may be different in updated versions.
Contents
Quick Start Guide
Downloading and Installing Slicer
|
General Introduction
Slicer Welcome Tutorial
|
Slicer4Minute Tutorial
|
3D Visualization
Slicer4 Data Loading and 3D Visualization
|
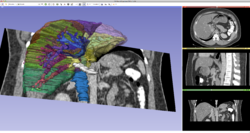
Slicer4 3D Visualization of DICOM images for Radiology Applications
|
Tutorials for software developers
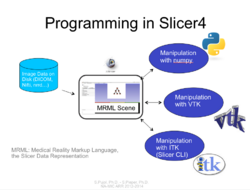
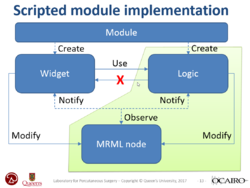
Slicer4 Programming Tutorial
|
PerkLab's Slicer bootcamp training materials
|
Slicer script repository
For additional Python scripts examples, please visit the Script Repository page
Developing and contributing extensions for 3D Slicer
|
Segmentation
Slicer4 Image Segmentation
|
|
|
|
|
|
Registration
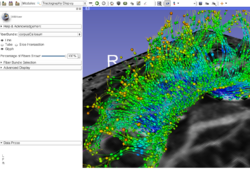
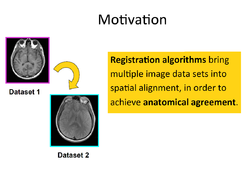
Slicer4 Image Registration
|
|
- Based on: 3D Slicer version 4.8
Slicer Registration Case Library
|
 |} |}
Slicer ExtensionsSlicer4 Diffusion Tensor Imaging Tutorial
Slicer4 Neurosurgical Planning Tutorial
Slicer4 Quantitative Imaging tutorial
Slicer4 IGT
Slicer4 Radiation Therapy Tutorial
Slicer Pathology
SPHARM-PDM
Fiber Bundle Volume Measurement
3D Slicer version 4.7 Tutorial ContestFor previous editions of the contest, please visit the 3D Slicer Tutorial Contests page Segmentation for 3D printing
Slicer Pathology
Simple Python Tool for Quality Control of DWI data
SPHARM-PDM
Integration of Robot Operating System (ROS) and 3D Slicer using OpenIGTLink
Fiber Bundle Volume Measurement
YouTube videosAdditional non-curated videos-based demonstrations using 3D Slicer are accessible on YouTube. Teams Contributions
International resourcesResources in Chinese
Resources in German
Murat Maga's blog posts about using 3D Slicer for biologyUsing the (legacy) EditorFast GrowCut
Use case: Slicer in paleontologyThis set of tutorials about the use of slicer in paleontology is very well written and provides step-by-step instructions. Even though it covers slicer version 3.4, many of the concepts and techniques have applicability to the new version and to any 3D imaging field:
|