Difference between revisions of "Documentation/Nightly/Extensions/SlicerHNSegmenter"
| Line 27: | Line 27: | ||
== Deployed model == | == Deployed model == | ||
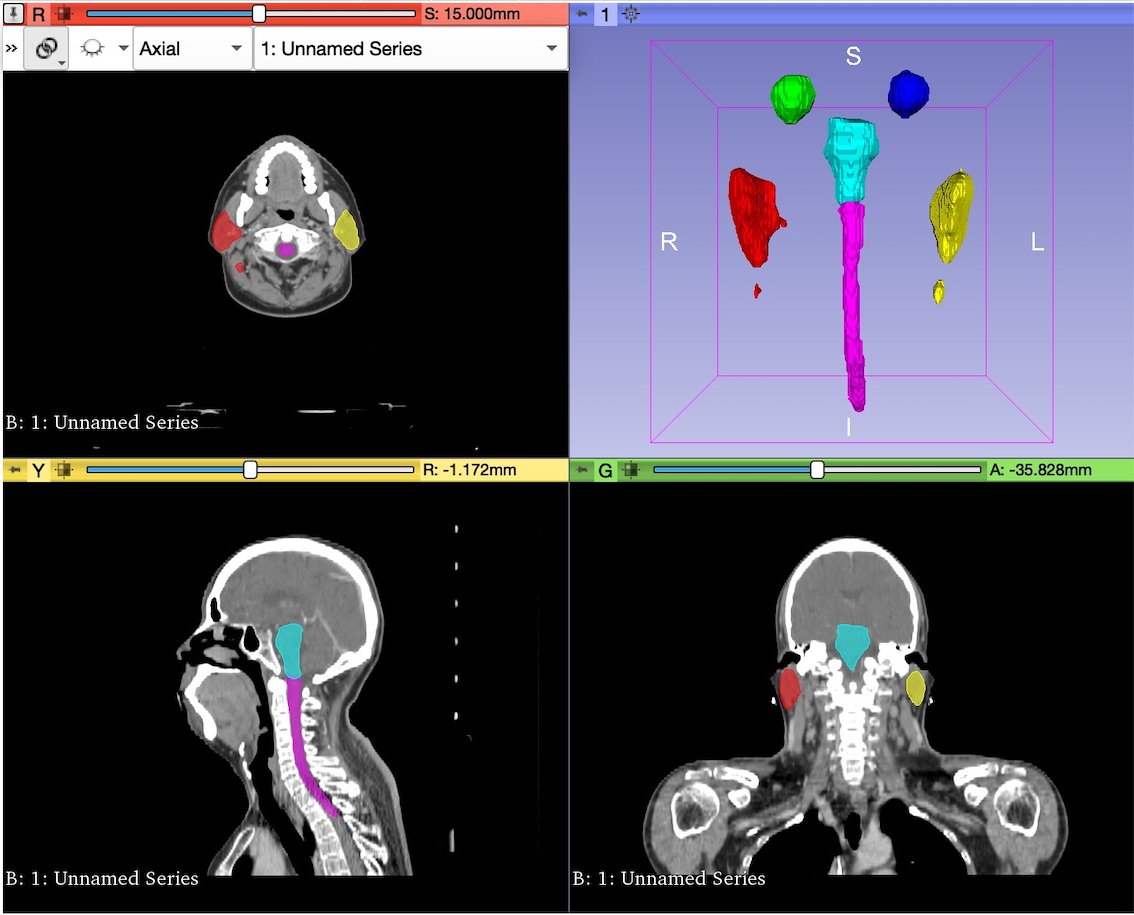
The dataset used to test and evaluate the performance of the module is composed by a set of CT scans obtained from the Structure Segmentation for Radiotherapy Planning Challenge 2019, a competition of MICCAI 2019 Challenge. Find [https://structseg2019.grand-challenge.org/Home/ here] the link. | The dataset used to test and evaluate the performance of the module is composed by a set of CT scans obtained from the Structure Segmentation for Radiotherapy Planning Challenge 2019, a competition of MICCAI 2019 Challenge. Find [https://structseg2019.grand-challenge.org/Home/ here] the link. | ||
| − | + | [[File:segmentationresults.jpg]] | |
| − | segmentationresults.jpg | ||
| − | |||
== Relevant Links == | == Relevant Links == | ||
[https://github.com/inesmunozz/Slicer-HN_Segmenter/tree/master Slicer HN-Segmenter Module Source Code] | [https://github.com/inesmunozz/Slicer-HN_Segmenter/tree/master Slicer HN-Segmenter Module Source Code] | ||
Revision as of 09:51, 22 September 2019
Home < Documentation < Nightly < Extensions < SlicerHNSegmenterContents
Module Description
HN Segmenter module is a 3D Slicer module intended to provide segmentations to a given CT head and neck image. It provides a user interface to connect with Docker and obtain 6 organs-at-risk (OARs) segmentations: left and right eye, left and right parotid, brainstem and spinal cord.
It constitutes a part of a project designed to accelerate the treatment planning in Radiotherapy by reducing planning time with automatic segmentation tools, instead of manual segmentations.
Setup Guide
In order to use HN Segmenter, Docker is required to be installed and configured properly.
Once installed, the Docker Image that stores the Neural Network should be pulled from the repository by typing the following code in the Terminal of your computer:
$docker pull inesmunoz/hn-segmenter
The pull will take few minutes to download, and it takes 2.69GB of memory.
Toc check if the Image was downloaded correctly one can type:
$docker images
And find the image there.
Now, the module is ready to execute the communications with Docker to obtain segmentations.
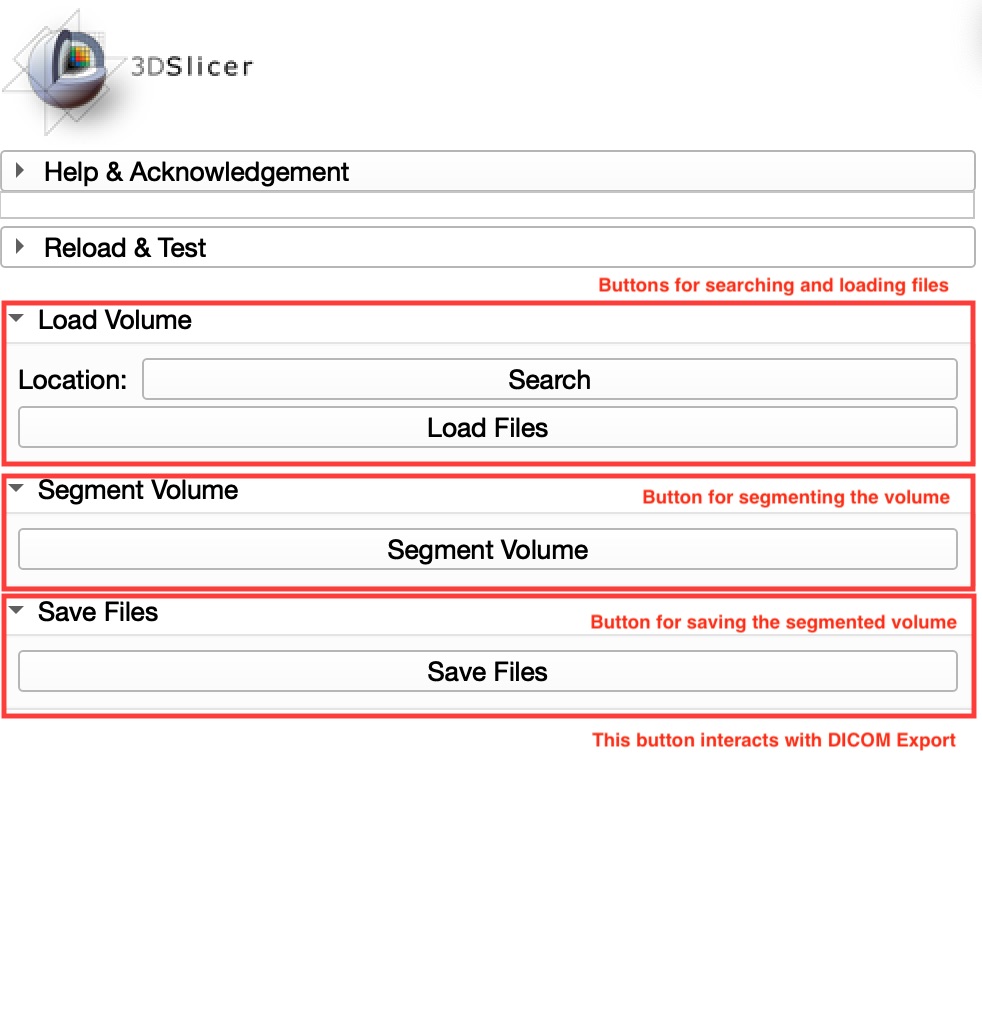
Panels and their use
The user interface is composed by only 3 sections, according to the three main functions of the module: loading an image, obtaining segmentations and saving the new volume.
This is possible thanks to the segmentation button, which performs few hidden functions. This button managed to automate the conversion of DICOM to nifty format, the importation of labelmaps into segmentations and all Terminal calls to execute Docker.
Deployed model
The dataset used to test and evaluate the performance of the module is composed by a set of CT scans obtained from the Structure Segmentation for Radiotherapy Planning Challenge 2019, a competition of MICCAI 2019 Challenge. Find here the link.