Difference between revisions of "Documentation/4.8/Training"
| Line 39: | Line 39: | ||
|} | |} | ||
| + | =3D Visualization= | ||
==Slicer4 Data Loading and 3D Visualization == | ==Slicer4 Data Loading and 3D Visualization == | ||
{|width="100%" | {|width="100%" | ||
| Line 52: | Line 53: | ||
|} | |} | ||
| − | = | + | ==Slicer4 3D Visualization of DICOM images for Radiology Applications== |
| + | {|width="100%" | ||
| + | | | ||
| + | *The [[Media:3DSlicer_Dicom_RSNA2015_SoniaPujol.pdf |3D Visualization of DICOM images for Radiology Applications]] course guides through 3D data loading and visualization of DICOM images for Radiology Applications in Slicer4. | ||
| + | *Author: Sonia Pujol, Ph.D., Kitt Shaffer, M.D., Ph.D., Ron Kikinis, M.D. | ||
| + | *Audience: Radiologists and users of Slicer who need a more comprehensive overview over Slicer4 visualization capabilities. | ||
| + | *Modules: DICOM, Volumes, Volume Rendering, Models. | ||
| + | *Based on: 3D Slicer version 4.5 | ||
| + | *The [[Media:3DVisualization_DICOM_images_part1.zip | 3DVisualizationDICOM_part1]] and [[Media:3DVisualization_DICOM_images_part2.zip | 3DVisualizationDICOM_part2]] datasets contain a series of MR and CT scans, and 3D models of the brain, lung and liver. | ||
| + | |align="right"| | ||
| + | [[Image:Slicer4RSNA_2.png|right|250px|]] | ||
| + | |} | ||
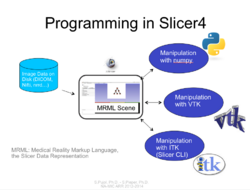
| + | =Programming= | ||
== Slicer4 Programming Tutorial == | == Slicer4 Programming Tutorial == | ||
| Line 79: | Line 92: | ||
|} | |} | ||
| − | = | + | =Segmentation= |
| + | == Slicer4 Image Segmentation == | ||
| + | {|width="100%" | ||
| + | | | ||
| + | * Segmentation for 3D printing: shows how to use the Segment Editor module for combining CAD designed parts with patient-specific models. | ||
| + | ** '''[https://discourse.slicer.org/t/new-video-tutorial-for-segment-editor-lumbar-spine-segmentation-for-3d-printing/700 Video tutorial]'''. Author: Hillary Lia. | ||
| + | ** '''[[Documentation/{{documentation/version}}/Training#Segmentation_for_3D_printing|Segmentation for 3D printing Step-by-step tutorial]]'''. Author: Csaba Pinter, MSc | ||
| + | ** Audience: Users and developers interested in segmentation and 3D printing | ||
| + | ** Based on: 3D Slicer version 4.7 | ||
| + | |align="right"|[[Image:20170717_3DPrintingTutorialYoutube.PNG|280px]] | ||
| + | |--- | ||
| + | | | ||
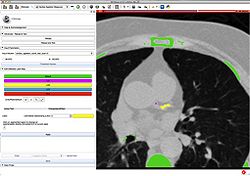
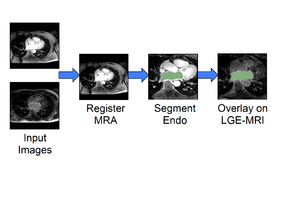
| + | * '''[https://youtu.be/BJoIexIvtGo Video tutorial: Whole heart segmentation from cardiac CT]''' shows how to use the Segment Editor module for segmenting heart ventricles, atria, and great vessels from cardiac CT volumes. | ||
| + | ** Author: Andras Lasso, PhD | ||
| + | ** Audience: Users who need to segment heart structures, for example for visualization, quantification, or simulation. | ||
| + | ** Sample data set: http://slicer.kitware.com/midas3/download/bitstream/738905/CTA-cardio2.nrrd | ||
| + | ** Based on: 3D Slicer version 4.8 | ||
| + | |align="right"|[[Image:WholeHeartSegYoutube.png|280px]] | ||
| + | |} | ||
| + | |||
| + | =Registration= | ||
| + | |||
| + | == Slicer4 Image Registration == | ||
| + | |||
| + | {|width="100%" | ||
| + | | | ||
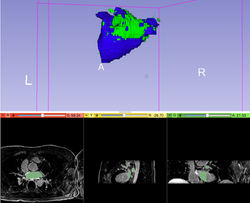
| + | *The [https://www.slicer.org/slicerWiki/index.php/File:RegistrationTutorial_3DSlicer4.5_spujol.pdf Registration tutorial] shows how to perform intra- and inter-subject registration within Slicer. | ||
| + | * Authors: Sonia Pujol, Ph.D., Dominik Meier, Ph.D., Ron Kikinis, M.D. | ||
| + | * Audience: Users and developers interested in image registration | ||
| + | * Dataset: [[Media:RegistrationData.zip| 3D Slicer Registration Data]] | ||
| + | |align="right"|[[File:registration_Slicer4.png|250px]] | ||
| + | |} | ||
| + | *Based on: 3D Slicer version 4.5 | ||
| + | See [[Documentation/{{documentation/version}}/Registration/RegistrationLibrary|the Registration Library for worked out registration examples with data]]. | ||
| + | |||
| + | == Slicer Registration Case Library== | ||
| + | {|width="100%" | ||
| + | | | ||
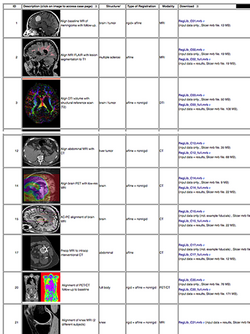
| + | *The ''[[Documentation/{{documentation/version}}/Registration/RegistrationLibrary|Slicer Registration Case Library]]'' provides many real-life example cases of using the Slicer registration tools. They include the dataset and step-by-step instructions to follow and try yourself. | ||
| + | :Author: Dominik Meier, Ph.D. | ||
| + | :Audience: users interested learning/applying Slicer image registration technology | ||
| + | |align="right"|[[Image:RegLib_table.png|250px|link=http://wiki.slicer.org/slicerWiki/index.php/Documentation/{{documentation/version}}/Registration/RegistrationLibrary]] | ||
| + | |} | ||
| + | |||
| + | =Slicer Extensions= | ||
==Slicer4 Diffusion Tensor Imaging Tutorial == | ==Slicer4 Diffusion Tensor Imaging Tutorial == | ||
{|width="100%" | {|width="100%" | ||
| Line 109: | Line 166: | ||
|} | |} | ||
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Slicer4 Quantitative Imaging tutorial== | ==Slicer4 Quantitative Imaging tutorial== | ||
| Line 148: | Line 194: | ||
|} | |} | ||
| − | == Slicer4 | + | |
| + | ==Slicer4 Radiation Therapy Tutorial == | ||
{|width="100%" | {|width="100%" | ||
| | | | ||
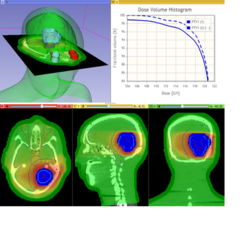
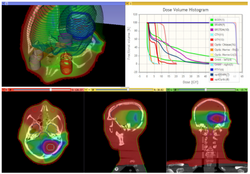
| − | * | + | * The [https://github.com/SlicerRt/SlicerRtDoc/raw/master/tutorials/SlicerRT_WorldCongress_TutorialIGRT.pdf SlicerRT tutorial] is an introduction to the Radiation Therapy functionalities of Slicer. |
| − | + | * Author: Csaba Pinter, Andras Lasso, An Wang, Gregory C. Sharp, David Jaffray, Gabor Fichtinger. | |
| − | + | * Dataset: [http://slicer.kitware.com/midas3/download/item/205404/SlicerRT_WorldCongress_TutorialIGRT_Dataset.zip download] from MIDAS server | |
| − | + | * Based on Slicer 4.7 | |
| − | + | |align="right"| | |
| − | + | <!-- [[Image:TUTORIAL-IMAGE-HERE.png|right|150px|]] --> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | * | ||
| − | |||
| − | |align="right"|[[Image: | ||
|} | |} | ||
| − | |||
| + | ==Slicer Pathology== | ||
{|width="100%" | {|width="100%" | ||
| | | | ||
| − | *The [ | + | *The [[Documentation/{{documentation/version}}/Extensions/SlicerPathology|Slicer Pathology Tutorial]] describes how to use the corresponding tools for automatic and semi-automatic pathology image segmentation. |
| − | * | + | *Author: Erich Bremer (Stonybrook), Andriy Fedorov (Brigham and Women’s Hospital) |
| − | + | *Dataset: Available directly with the Slicer Pathology Slicer extension. | |
| − | * Dataset: | + | |align="right"| |
| − | |align="right"|[[File: | + | [[File:SlicerPathologyScreenShot8.png | 200px]]. |
|} | |} | ||
| − | |||
| − | |||
| − | == | + | ==SPHARM-PDM== |
| + | {|width="100%" | ||
| + | | | ||
| + | *The [https://www.nitrc.org/docman/view.php/308/1982/SPHARM-PDM_Tutorial_July2015.pdf SPHARM-PDM Tutorial] describes how to use SPHARM-PDM and ShapePopulationViewer Slicer extensions to respectively compute point-based models using a parametric boundary description for the computing of Shape Analysis and perform the quality control between the different models. | ||
| + | *Author: Jonathan Perdomo (UNC), Beatriz Paniagua (Kitware Inc.) | ||
| + | *Dataset: [https://www.nitrc.org/docman/view.php/308/1981/SPHARM_Tutorial_Data_July2015.zip Tutorial Data] | ||
| + | |align="right"| | ||
| + | [[File:SlicerWinterProjectWeek2017-SPHARM-PDM.png | 200px]]. | ||
| + | |} | ||
| + | ===Fiber Bundle Volume Measurement=== | ||
{|width="100%" | {|width="100%" | ||
| | | | ||
| − | * The [ | + | *The [http://www.na-mic.org/Wiki/images/5/57/Fiber_Bundle_Volume_Measurement.pptx Fiber Bundle Volume Measurement Tutorial] aim is to calculate the volume of the fiber bundle that passes through the Corpus Callosum(CC). Following this tutorial, you’ll be able to (1) convert fiber bundles to label map and (2) calculate volume measurements from the fiber bundles. |
| − | * Author: | + | *Author: Shun Gong (Shanghai Changzheng Hospital, China) |
| − | * Dataset: [http:// | + | *Dataset: [http://www.na-mic.org/Wiki/images/4/4c/FiberVolume_data.zip Tutorial data]: The following data are provided: Baseline image, Down sampled whole brain tractography (conducted as in the [[Documentation/{{documentation/version}}/Training#Slicer4_Diffusion_Tensor_Imaging_Tutorial|DWI tutorial]] and down-sampled to about 10000 fibers using Tractography Display module), Corpus callosum label map (drawn as in the [[Documentation/{{documentation/version}}/Training#Slicer4_Diffusion_Tensor_Imaging_Tutorial|DWI tutorial]]). |
| − | |||
|align="right"| | |align="right"| | ||
| − | + | [[File:SlicerWinterProjectWeek2017-FiberBundleVolumeMeasurements.png | 200px]]. | |
|} | |} | ||
| − | |||
| − | |||
| − | |||
| − | |||
= 3D Slicer Tutorial contests= | = 3D Slicer Tutorial contests= | ||
| Line 416: | Line 457: | ||
|} | |} | ||
| − | = | + | =Teams Contributions = |
{|width="100%" | {|width="100%" | ||
| | | | ||
| Line 439: | Line 480: | ||
---- | ---- | ||
<br> | <br> | ||
| − | + | See the collection of videos on the [http://vimeo.com/album/2363361 Kitware vimeo album]. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
= External Resources = | = External Resources = | ||
| − | + | ==Non-curated videos== | |
| − | == Resources | + | Additional non-curated videos-based demonstrations using 3D Slicer are accessible on [http://www.youtube.com/results?search_query=3d+slicer&sm=3 You Tube]. |
| − | + | == Resources in Chinese == | |
| − | |||
{|width="100%" | {|width="100%" | ||
| | | | ||
| Line 505: | Line 539: | ||
* [http://openpaleo.blogspot.com/2009/03/3d-slicer-tutorial-part-v.html Open Source Paleontologist: 3D Slicer: The Tutorial Part V] | * [http://openpaleo.blogspot.com/2009/03/3d-slicer-tutorial-part-v.html Open Source Paleontologist: 3D Slicer: The Tutorial Part V] | ||
* [http://openpaleo.blogspot.com/2009/03/3d-slicer-tutorial-part-vi.html Open Source Paleontologist: 3D Slicer: The Tutorial Part VI] | * [http://openpaleo.blogspot.com/2009/03/3d-slicer-tutorial-part-vi.html Open Source Paleontologist: 3D Slicer: The Tutorial Part VI] | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Revision as of 14:46, 11 April 2018
Home < Documentation < 4.8 < Training
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction: Slicer 4.8 Tutorials
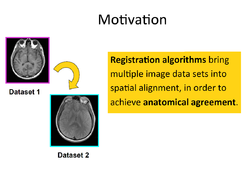
- This page contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment (version 4.8 release) to accomplish certain tasks.
- For tutorials for other versions of Slicer, please visit the Slicer training portal.
- For "reference manual" style documentation, please visit the Slicer 4.8 documentation page
- For questions related to the Slicer4 Training Compendium, please send an e-mail to Sonia Pujol, Ph.D., Director of Training of 3D Slicer.
- Some of these tutorials are based on older releases of 3D Slicer. The concepts are still useful but bear in mind that some interface elements and features will be different in updated versions.
Contents
- 1 Introduction: Slicer 4.8 Tutorials
- 2 General Introduction
- 3 3D Visualization
- 4 Programming
- 5 Segmentation
- 6 Registration
- 7 Slicer Extensions
- 8 3D Slicer Tutorial contests
- 9 Teams Contributions
- 10 External Resources
General Introduction
Slicer Welcome Tutorial
|
Slicer4Minute Tutorial
|
3D Visualization
Slicer4 Data Loading and 3D Visualization
|
Slicer4 3D Visualization of DICOM images for Radiology Applications
|
Programming
Slicer4 Programming Tutorial
|
For additional Python scripts examples, please visit the Script Repository page
Developing and contributing extensions for 3D Slicer
|
Segmentation
Slicer4 Image Segmentation
|
|
|
|
Registration
Slicer4 Image Registration
|
|
- Based on: 3D Slicer version 4.5
See the Registration Library for worked out registration examples with data.
Slicer Registration Case Library
|
|
Slicer Extensions
Slicer4 Diffusion Tensor Imaging Tutorial
|
Slicer4 Neurosurgical Planning Tutorial
|
Slicer4 Quantitative Imaging tutorial
|
Slicer4 IGT
|
Slicer4 Radiation Therapy Tutorial
|
Slicer Pathology
|
SPHARM-PDM
|
Fiber Bundle Volume Measurement
|
3D Slicer Tutorial contests
Winter 2017 Tutorial contest
Segmentation for 3D printing
|
Slicer Pathology
|
Simple Python Tool for Quality Control of DWI data
|
SPHARM-PDM
|
Integration of Robot Operating System (ROS) and 3D Slicer using OpenIGTLink
|
Fiber Bundle Volume Measurement
|
Winter 2016 Tutorial contest
Subject Hierarchy
|
Fiber Bundle Selection and Scalar Measurements
|
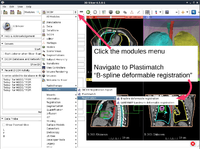
Plastimatch
|
UKF
|
Summer 2014 Tutorial contest
Cardiac Agatston Tutorial
|
CMR Toolkit LA workflow
|
Summer 2013 Tutorial contest
Cardiac MRI Toolkit
|
HelloCLI
|
SlicerRT
|
DTIPrep
|
Summer 2012 Tutorial contest
Automatic Left Atrial Scar Segmenter
|
Qualitative and quantitative comparison of two RT dose distributions
|
Dose accumulation for adaptive radiation therapy
|
WebGL Export
|
OpenIGTLink
|
Teams Contributions
|
|
|
|
See the collection of videos on the Kitware vimeo album.
External Resources
Non-curated videos
Additional non-curated videos-based demonstrations using 3D Slicer are accessible on You Tube.
Resources in Chinese
|
A 3D Slicer community on WeChat in China offers many tutorials and clinical examples in Chinese. Note that the images are of interest to non-Chinese speakers and Google Translate does a reasonable job of translating some of the text. |
Resources in German
- A series of four YouTube videos on python programming in Slicer (German narration with English subtitles)
Murat Maga's blog posts about using 3D Slicer for biology
Using the (legacy) Editor
Fast GrowCut
|

|
Use case: Slicer in paleontology
This set of tutorials about the use of slicer in paleontology is very well written and provides step-by-step instructions. Even though it covers slicer version 3.4, many of the concepts and techniques have applicability to the new version and to any 3D imaging field:
- Open Source Paleontologist: 3D Slicer: The Tutorial
- Open Source Paleontologist: 3D Slicer: The Tutorial Part II
- Open Source Paleontologist: 3D Slicer: The Tutorial Part III
- Open Source Paleontologist: 3D Slicer: The Tutorial Part IV
- Open Source Paleontologist: 3D Slicer: The Tutorial Part V
- Open Source Paleontologist: 3D Slicer: The Tutorial Part VI