Difference between revisions of "Announcements:Slicer3.2"
| Line 19: | Line 19: | ||
|} | |} | ||
| − | + | =Overview= | |
| − | <gallery caption="Slicer v3.2 - New and Improved Feature Highlights" widths=" | + | <gallery caption="Slicer v3.2 - New and Improved Feature Highlights" widths="250px" heights="150px" perrow="4"> |
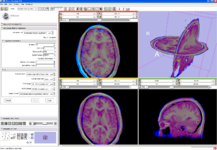
| − | Image:VolRend.png|[[Modules:VolumeRendering-Documentation|Fully integrated volume rendering]] with cropping | + | Image:VolRend.png|Volume Rendering: [[Modules:VolumeRendering-Documentation|Fully integrated volume rendering]] with cropping for easy exploration of volumetric data |
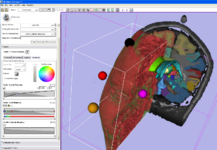
| − | Image:ImplicitPlane.png|Implicit Slice Widget for specifying oblique views (part of the [http://www.vtk.org/Wiki/VTK_Widget_Examples VTK widget family]). | + | Image:ImplicitPlane.png|Implicit Slice Widget: An interactive tool for specifying oblique views (part of the [http://www.vtk.org/Wiki/VTK_Widget_Examples VTK widget family]). |
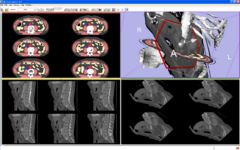
| − | Image:LightBox.png|[[Slicer3:Tools_for_Radiologists|The Lightbox]]: | + | Image:LightBox.png|[[Slicer3:Tools_for_Radiologists|The Lightbox]]: Functionality for displaying cross-sectional data in columns and rows. |
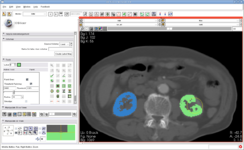
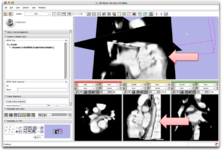
| − | Image:Editor-v3-2.png|[[Modules:Editor-Documentation|Interactive Editor]] allows interactive segmentation with | + | Image:Editor-v3-2.png|[[Modules:Editor-Documentation|Interactive Editor:]] This new module allows interactive segmentation with robust 2D and 3D algorithms |
| − | Image:IO.png|[[Documentation|IO capabilities]] include DICOM, NRRD, NIFTI, Tiff, JPG, Freesurfer and | + | Image:IO.png|IO: [[Documentation|Improved IO capabilities]] include support for DICOM, NRRD, NIFTI, Tiff, JPG, Freesurfer, FITS and a number of other formats |
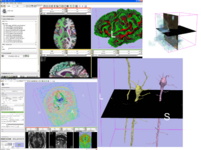
Image:Line-glyph-tracts.jpg|Glyphs on tracts:<br>[[Modules:DTMRI-Documentation|New Diffusion Imaging infrastructure]] includes<br>dicom import<br>gradient editor<br>visualiztion | Image:Line-glyph-tracts.jpg|Glyphs on tracts:<br>[[Modules:DTMRI-Documentation|New Diffusion Imaging infrastructure]] includes<br>dicom import<br>gradient editor<br>visualiztion | ||
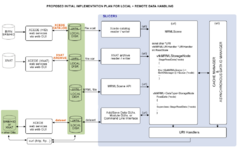
Image:DataLoadingStartPlan.png|[[Slicer3:Remote_Data_Handling|Remote Data Handling]] allows uploads and downloads from image informatics frameworks such as BIRN, and XNAT | Image:DataLoadingStartPlan.png|[[Slicer3:Remote_Data_Handling|Remote Data Handling]] allows uploads and downloads from image informatics frameworks such as BIRN, and XNAT | ||
Revision as of 17:28, 27 May 2008
Home < Announcements:Slicer3.2Slicer 3.2
|
Slicer 3.2, a major release of the Slicer software platform, is scheduled for release on Friday, May 30, 2008. The new release contains hundreds of changes to the software. Highlights include:
Click here to download different versions of Slicer3 and find pointers to the source code, mailing lists and bug tracker. Please note that Slicer continues to be a research package and is not intended for clinical use. Testing of functionality is an ongoing activity with high priority, however, some features of Slicer3 are not fully tested. |
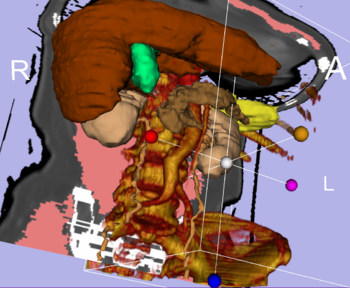
Integrated Volume Rendering: View of the abdominal atlas Bone and large vessels are volume rendered. |
Overview
- Slicer v3.2 - New and Improved Feature Highlights
Volume Rendering: Fully integrated volume rendering with cropping for easy exploration of volumetric data
Implicit Slice Widget: An interactive tool for specifying oblique views (part of the VTK widget family).
The Lightbox: Functionality for displaying cross-sectional data in columns and rows.
Interactive Editor: This new module allows interactive segmentation with robust 2D and 3D algorithms
IO: Improved IO capabilities include support for DICOM, NRRD, NIFTI, Tiff, JPG, Freesurfer, FITS and a number of other formats
Glyphs on tracts:
New Diffusion Imaging infrastructure includes
dicom import
gradient editor
visualiztionRemote Data Handling allows uploads and downloads from image informatics frameworks such as BIRN, and XNAT
OpenIGTLink interfaces to devices such as scanners, trackers, and robots
- Loadable Modules
- EM segementer
- Scenesnapshots