Difference between revisions of "Documentation/Nightly/Extensions/SlicerPathology"
m |
|||
| Line 31: | Line 31: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Tutorials}} | {{documentation/{{documentation/version}}/module-section|Tutorials}} | ||
| − | <embedvideo service="youtube">https://www.youtube.com/watch?v= | + | <embedvideo service="youtube">https://www.youtube.com/watch?v=n6RtJoU9nGQ</embedvideo> |
{| | {| | ||
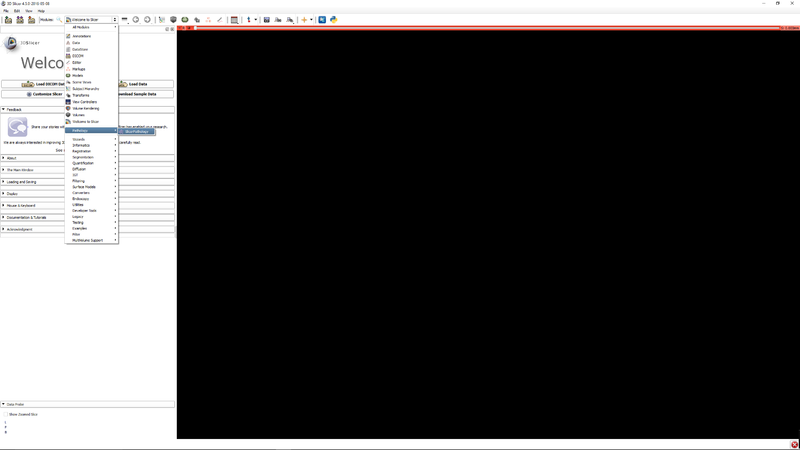
|[[File:SlicerPathologyScreenShot2.png|thumb|800px|Step 1 - Go to the SlicerPathology Extension.]] | |[[File:SlicerPathologyScreenShot2.png|thumb|800px|Step 1 - Go to the SlicerPathology Extension.]] | ||
Latest revision as of 20:54, 7 December 2016
Home < Documentation < Nightly < Extensions < SlicerPathology
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: SlicerPathology Contributor 1: Yi Gao |
Module Description
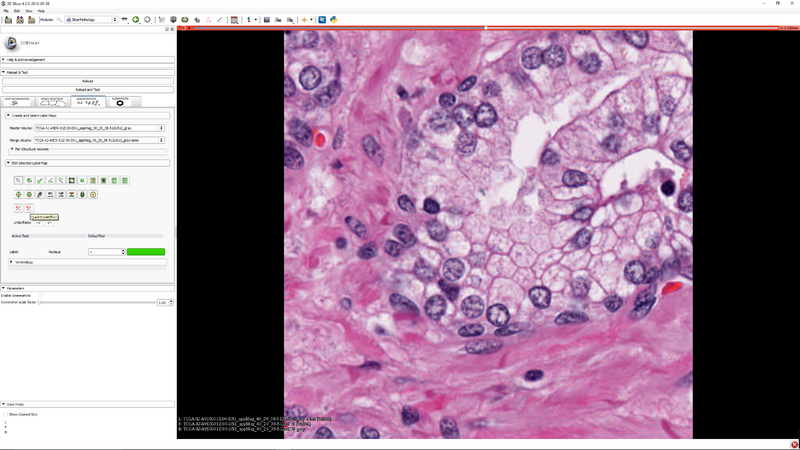
This extension provides tools for automatic and semi-automatic pathology image segmentation.
Use Cases
| Section under construction. |
Tutorials
References
- Quantitative Image Informatics for Cancer Research (QIICR)
- Open Source Computer Vision
- The Cancer Genome Atlas
Information for Developers
The source code for SlicerPathology is available at https://github.com/SBU-BMI/SlicerPathology