|
|
| Line 58: |
Line 58: |
| | | | |
| | The first 3D Slicer training events using Slicer 4.5 will be organized at [http://www.na-mic.org/Wiki/index.php/MGH_2015 Massachusetts General Hospital (MGH), Boston, MA], [http://www.na-mic.org/Wiki/index.php/Brown_2015 Brown University, Providence, RI] and [http://www.na-mic.org/Wiki/index.php/RSNA_2015 RSNA 2015, Chicago, Il]. | | The first 3D Slicer training events using Slicer 4.5 will be organized at [http://www.na-mic.org/Wiki/index.php/MGH_2015 Massachusetts General Hospital (MGH), Boston, MA], [http://www.na-mic.org/Wiki/index.php/Brown_2015 Brown University, Providence, RI] and [http://www.na-mic.org/Wiki/index.php/RSNA_2015 RSNA 2015, Chicago, Il]. |
| − |
| |
| − | <!--
| |
| − | * [https://www.youtube.com/channel/UC11x1iQ7ydSIFYw4L6wveXg?view_as=public 3D Slicer YouTube channel] has been reorganized, new videos developed by the 3D Slicer community added to the channel
| |
| − | * New [https://youtu.be/GGgP89uTOLo 3D Slicer Tutorial: How to Segment a Lumbar Vertebrae] has been added
| |
| − | * New addition to the [http://wiki.na-mic.org/Wiki/images/b/b2/DiffusionMRIanalysis_Tutorial_SoniaPujol_UNC2015.pdf DTI tutorial] with brain masking section and new DWI dataset
| |
| − | * New [http://www.slicer.org/slicerWiki/index.php/Documentation/{{documentation/version}}/Training#Slicer4_Image_Registration 3D Slicer Registration Tutorial] has been added to the compendium.
| |
| − | -->
| |
| − | <!--
| |
| − | Sonia: Is the link the latest one ?
| |
| − | * New dataset has been added to the [http://www.slicer.org/slicerWiki/images/1/1f/DiffusionMRIanalysis_Tutorial_SoniaPujol_2013.pdf 3D Slicer Diffusion Tensor Imaging Tutorial]
| |
| − | -->
| |
| | | | |
| | <gallery caption="New Tutorials" widths="250px" heights="150px"> | | <gallery caption="New Tutorials" widths="250px" heights="150px"> |
| | | | |
| | Image:SlicerYouTube.png| [https://www.youtube.com/channel/UC11x1iQ7ydSIFYw4L6wveXg?view_as=public 3D Slicer YouTube channel] has been reorganized, new videos developed by the 3D Slicer community added to the channel {{new}} | | Image:SlicerYouTube.png| [https://www.youtube.com/channel/UC11x1iQ7ydSIFYw4L6wveXg?view_as=public 3D Slicer YouTube channel] has been reorganized, new videos developed by the 3D Slicer community added to the channel {{new}} |
| − |
| |
| − | Image:RegistrationTutorial.png|New [http://www.slicer.org/slicerWiki/index.php/Documentation/{{documentation/version}}/Training#Slicer4_Image_Registration 3D Slicer Registration Tutorial] has been added to the compendium. {{new}}
| |
| − |
| |
| − | Image:VertebraTutorial.png|Video Tutorial: [https://youtu.be/GGgP89uTOLo How to Segment a Lumbar Vertebrae] {{new}}
| |
| − |
| |
| − | Image:2015 DTITutorial Addition of Brain Masking.png|New addition to the [http://wiki.na-mic.org/Wiki/images/b/b2/DiffusionMRIanalysis_Tutorial_SoniaPujol_UNC2015.pdf DTI tutorial] with brain masking section and new DWI dataset. {{updated}}
| |
| | | | |
| | | | |
| Line 90: |
Line 73: |
| | | | |
| | Image:AnglePlanes Logo.png|[[Documentation/{{documentation/version}}/Extensions/AnglePlanes|AnglePlanes]] This Module is used to calculate the angle between two planes by using the normals {{new}} | | Image:AnglePlanes Logo.png|[[Documentation/{{documentation/version}}/Extensions/AnglePlanes|AnglePlanes]] This Module is used to calculate the angle between two planes by using the normals {{new}} |
| − |
| |
| − | Image:Chest_Imaging_Platform.png|[http://www.chestimagingplatform.org/ Chest_Imaging_Platform] Chest Imaging Platform is an extension for quantitative CT imaging biomarkers for lung diseases {{new}}
| |
| − |
| |
| − | Image:DebuggingTools.png|[[Documentation/{{documentation/version}}/Extensions/DebuggingTools|DebuggingTools]] This extension contains various tools useful for developing and debugging modules {{new}}
| |
| − |
| |
| − | Image:SlicerExtension-DeveloperToolsForExtensions.png|[[Documentation/{{documentation/version}}/Extensions/DeveloperToolsForExtensions|DeveloperToolsForExtensions]] This extension offers different tools to help developers when they create Slicer extension {{new}}
| |
| − |
| |
| − | Image:DTI-Reg.png|[[Documentation/{{documentation/version}}/Extensions/DTI-Reg|DTI-Reg]] DTI-Reg is an extension that performs pair-wise DTI registration, using scalar FA map to drive the registration {{new}}
| |
| − |
| |
| − | Image:EasyClipLogo.png|[[Documentation/{{documentation/version}}/Extensions/EasyClip|EasyClip]] This Module is used to clip one or different 3D Models according to a predetermined plane {{new}}
| |
| − |
| |
| − | Image:GraphCutSegment.png|[http://publish.uwo.ca/~dchen285/GraphCutSegment/GraphCutSegment.html GraphCutSegment] This is a segment extension using graph cut and star shape algorithm {{new}}
| |
| − |
| |
| − | Image:LumpNav.png|[https://github.com/SlicerIGT/LumpNav LumpNav] Breast tumor resection using tracked ultrasound and cautery {{new}}
| |
| − |
| |
| − | Image:MarginCalculator Logo2 128.png|[[Documentation/{{documentation/version}}/Extensions/MarginCalculator|MarginCalculator]] The Matlab Bridge extension allows running Matlab scripts as command-line interface (CLI) modules directly from 3D Slicer {{new}}
| |
| − |
| |
| − | Image:MeshStatisticsExtension.png|[[Documentation/{{documentation/version}}/Extensions/MeshStatistics|MeshStatistics]] Mesh Statistics allows users to compute descriptive statistics over specific and predefined regions {{new}}
| |
| − |
| |
| − | Image:Icon1.png|[[Documentation/{{documentation/version}}/Extensions/MeshToLabelMap|MeshToLabelMap]] This extension computes a label map from a 3D model {{new}}
| |
| − |
| |
| − | Image:PET-IndiC.png|[[Documentation/{{documentation/version}}/Extensions/PET-IndiC|PET-IndiC]] The PET-IndiC Extension allows for fast segmentation of regions of interest and calculation of quantitative indices {{new}}
| |
| − |
| |
| − | Image:DPetBrainQuantification.png|[http://gti-fing.github.io/SlicerPetSpectAnalysis/ PetSpectAnalysis] First Version of the Pet Spect Analysis Extension {{new}}
| |
| − |
| |
| − | Image:PETTumorSegmentationExtensionIcon.png|[[Documentation/{{documentation/version}}/Extensions/PETTumorSegmentation|PETTumorSegmentation]] Tumor and lymph node segmentation in PET scans with a specialized Editor effect {{new}}
| |
| − |
| |
| − | Image:PickAndPaintExtension.png|[[Documentation/{{documentation/version}}/Extensions/PickAndPaint|PickAndPaint]] Pick 'n Paint tool allows users to select ROIs on a reference model and to propagate it over different time point models {{new}}
| |
| − |
| |
| − | Image:Q3DC.png|[[Documentation/{{documentation/version}}/Extensions/Q3DC|Q3DC]] This extension contains one module of the same name {{new}}
| |
| − |
| |
| − | Image:ResampleDTIlogEuclidean-128x128-icon.png|[[Documentation/{{documentation/version}}/Extensions/ResampleDTIlogEuclidean|ResampleDTIlogEuclidean]] This resamples Diffusion Tensor Images (DTI) in the log-euclidean framework {{new}}
| |
| − |
| |
| − | Image:SlicerHeart Logo 128x128.png|[[Documentation/{{documentation/version}}/Extensions/SlicerHeart|SlicerHeart]] Modules for cardiac analysis and intervention planning and guidance {{new}}
| |
| − |
| |
| − | Image:SlicerProstate Logo 1.0 128x128.png|[[Documentation/{{documentation/version}}/Extensions/SlicerProstate|SlicerProstate]] SlicerProstate extension hosts various modules to facilitate processing and management of prostate image data, utilizing prostate images in image-guided interventions and development of the imaging biomarkers of the prostate cancer {{new}}
| |
| − |
| |
| − | Image:Slicer-Wasp.png|[https://github.com/Tomnl/Slicer-Wasp Slicer-Wasp] A module to perform a series of ITK watershed segmentation (without seeds) and then let the user create a label map out of selected components {{new}}
| |
| − |
| |
| − | Image:T1 Mapping Logo Resized.png|[[Documentation/{{documentation/version}}/Extensions/T1Mapping|T1Mapping]] T1 mapping estimates effective tissue parameter maps (T1) from multi-spectral FLASH MRI scans with different flip angles {{new}}
| |
| | | | |
| | <!-- You could user either {{new}} or {{updated}} macros. --> | | <!-- You could user either {{new}} or {{updated}} macros. --> |
| Line 136: |
Line 79: |
| | == Improved Extensions in Slicer 4.5 == | | == Improved Extensions in Slicer 4.5 == |
| | | | |
| − | * [http://www.nitrc.org/projects/abc ABC]
| |
| | * [[Documentation/{{documentation/version}}/Extensions/CMFreg|CMFreg]] | | * [[Documentation/{{documentation/version}}/Extensions/CMFreg|CMFreg]] |
| − | * [[Documentation/{{documentation/version}}/Extensions/CurveMaker|CurveMaker]]
| + | <!-- Add entry here --> |
| − | * [[Documentation/{{documentation/version}}/Extensions/DTIProcess|DTIProcess]]
| |
| − | * [[Documentation/{{documentation/version}}/Modules/GelDosimetry|GelDosimetryAnalysis]]
| |
| − | * [[Documentation/{{documentation/version}}/Extensions/IntensitySegmenter|IntensitySegmenter]]
| |
| − | * [[Documentation/{{documentation/version}}/Extensions/MatlabBridge|MatlabBridge]]
| |
| − | * [[Documentation/{{documentation/version}}/Extensions/ModelToModelDistance|ModelToModelDistance]]
| |
| − | * [[Documentation/{{documentation/version}}/Extensions/NeedleFinder|NeedleFinder]]
| |
| − | * [[Documentation/{{documentation/version}}/Extensions/PETDICOM|PETDICOMExtension]]
| |
| − | * [[Documentation/{{documentation/version}}/Extensions/PercutaneousApproachAnalysis|PercutaneousApproachAnalysis]]
| |
| − | * [[Documentation/{{documentation/version}}/Extensions/Reporting|Reporting]]
| |
| − | * [[Documentation/{{documentation/version}}/Extensions/SlicerIGT|SlicerIGT]]
| |
| − | * [[Documentation/{{documentation/version}}/Extensions/SlicerRT|SlicerRT]]
| |
| − | * [[Documentation/{{documentation/version}}/Extensions/VolumeClip|VolumeClip]]
| |
| − | * [[Documentation/{{documentation/version}}/Extensions/TCIABrowser|TCIABrowser]]
| |
| | | | |
| | == Extensions removed from Slicer 4.5 == | | == Extensions removed from Slicer 4.5 == |
| | | | |
| − | * houghTransformCLI: Removed by the original author because it was not needed anymore. | + | <!-- * houghTransformCLI: Removed by the original author because it was not needed anymore. --> |
| | | | |
| | == Extensions renamed == | | == Extensions renamed == |
| | | | |
| | + | <!-- |
| | * PyDevRemoteDebug -> [[Documentation/{{documentation/version}}/Extensions/DebuggingTools|DebuggingTools]] | | * PyDevRemoteDebug -> [[Documentation/{{documentation/version}}/Extensions/DebuggingTools|DebuggingTools]] |
| | * MultidimData -> [[Documentation/{{documentation/version}}/Extensions/Sequences|Sequences]] | | * MultidimData -> [[Documentation/{{documentation/version}}/Extensions/Sequences|Sequences]] |
| | * TrackerStabilizer -> [[Documentation/{{documentation/version}}/Extensions/TrackerStabilizer|Slicer-TrackerStabilizer]] | | * TrackerStabilizer -> [[Documentation/{{documentation/version}}/Extensions/TrackerStabilizer|Slicer-TrackerStabilizer]] |
| | * AirwaySegmentation -> [[Documentation/{{documentation/version}}/Extensions/AirwaySegmentation|Slicer-AirwaySegmentation]] | | * AirwaySegmentation -> [[Documentation/{{documentation/version}}/Extensions/AirwaySegmentation|Slicer-AirwaySegmentation]] |
| | + | --> |
| | | | |
| | = Other Improvements, Additions & Documentation = | | = Other Improvements, Additions & Documentation = |
| | | | |
| | == Optimization == | | == Optimization == |
| − | * Added support for loading of large images on windows.
| |
| − | * Reduced the memory footprint by fixing memory leaks
| |
| − | * Sped up by 2x slice rendering when the volume is linearly transformed.
| |
| − | * Improved reslicing speed of quasi-axis-aligned slices.
| |
| − | * Sped up annotation ROI display about 10x faster (100ms to 10ms).
| |
| − | * Improved tractography display performance.
| |
| − | * Prevent viewers flickering when only few nodes are removed.
| |
| − | * Improved performance of transform widget.
| |
| | | | |
| | == Transforms == | | == Transforms == |
| − | * Added support for copy/paste of linear transforms.
| |
| − | * Support saving of any transform to a grid transform.
| |
| − | * Addressed translation slider instability in rotate first mode.
| |
| − | * Improved composite transform support.
| |
| − | * Added support for thin plate spline transforms.
| |
| − | * Changed default ITK bspline transform writing file format to ITKv4.
| |
| − | * Addressed display issue for slice intersection display of non-linearly transformed model.
| |
| − | * Made transforms module translate-rotate first icon nicer.
| |
| | | | |
| | == DICOM == | | == DICOM == |
| − | * Improved DICOM browser adding a right click menu allowing delting and exporting to disk patients/studies/series
| |
| − | * Improved support for loading of DICOM referenced datasets.
| |
| − | * Added support for DICOM private dictionary
| |
| − | * Read and apply window/level found in DICOM volumes.
| |
| − | * Added support for Philips DWI datasets
| |
| − | * Fixed loading of DICOM image from files containing special characters.
| |
| | | | |
| | == Data processing == | | == Data processing == |
| − | * Added support for saving and loading of complex processing chain of CLI modules [http://www.na-mic.org/Bug/view.php?id=3240 #3240] [http://www.na-mic.org/Bug/view.php?id=3468 #3468]
| |
| | | | |
| | == CLI == | | == CLI == |
| − | * Added support for transfer of fiducial file to/from CLI modules.
| |
| − | * Added support for transfer of Hierarchy sub-scene to/from CLI modules.
| |
| − | * Fixed loading multivolumes produced by CLI modules.
| |
| | | | |
| | == Usability == | | == Usability == |
| − | * Built-in module categories and extensions categories are listed separately in module selector.
| |
| − | * Allow multi-selection in subject hierarchy.
| |
| − | * Generate high-resolution magnified screenshots for 3D view.
| |
| | | | |
| | == SubjectHierarchy == | | == SubjectHierarchy == |
| − | * Highlight nodes in subject hierarchy referenced by DICOM
| |
| − | * Added Transforms subject hierarchy plugin
| |
| − | * Added new [[Documentation/Labs/DICOMExport|DICOM export feature and scalar volume plugin]]. [http://www.na-mic.org/Bug/view.php?id=3163 #3163]
| |
| − | * Added generic folder plugin to subject hierarchy
| |
| − | * Subject hierarchy nodes automatically created for supported types
| |
| − | * Added auto-delete subject hierarchy children to Application Settings
| |
| − | * Added auto-create subject hierarchy option to Application Settings
| |
| − | * Subject hierarchy tree headers show icons instead of text
| |
| − | * Fixed subject hierarchy nodes creation when importing old scene [http://www.na-mic.org/Bug/view.php?id=3902 #3902]
| |
| − | * Fixed volume visibility controls. [http://www.na-mic.org/Bug/view.php?id=3893 #3893]
| |
| | | | |
| | == Python scripting == | | == Python scripting == |
| − | * Upgraded from Numpy 1.4.1 to NumPy 1.9.2
| |
| − | * Upgraded from Python 2.7.3 to Python 2.7.10
| |
| − | * Bundled nose python module into the Slicer package.
| |
| − | * Added logging functions for Python.
| |
| − | * Fixed VTKObservationMixin and added [[Documentation/Nightly/Developers/Python_scripting#How_can_I_access_callData_argument_in_a_VTK_object_observer_callback_function|documentation]].
| |
| − | * Updated IOManager and DICOMWidgets to emit <code>newFileLoaded()</code>.
| |
| − | * Added logging of Python console user input.
| |
| − | * Added accessor for vertex points of models as numpy array.
| |
| − | * Allow displayable manager to be introspected using python.
| |
| | | | |
| | == Editor == | | == Editor == |
| − | * Simplify UI using node selector for merge volume. [http://www.na-mic.org/Bug/view.php?id=2958 #2958]
| |
| − | * Warn user when GrowCut effect is attempted with unsupported image type.
| |
| − | * Added ability to delete only the selected structure.
| |
| − | * Ensure label map is updated each time a master volume is selected.
| |
| − | * Improved threshold paint effect to have inclusive ranges [http://www.na-mic.org/Bug/view.php?id=4066 #4066]
| |
| − | * Remember paint radius when effect are changed [http://www.na-mic.org/Bug/view.php?id=1853 #1853]
| |
| − | * Reset editor interface when scene is closed.
| |
| − | * Ensure a related merge volume is selected after a structure deleted.
| |
| | | | |
| | == Markups == | | == Markups == |
| − | * Added measurements between fiducials in right click menu [http://www.na-mic.org/Bug/view.php?id=1898 #1898]
| |
| − | * Added ability to refocus 3d cameras on a fiducial. [http://www.na-mic.org/Bug/view.php?id=3683 #3683]
| |
| | | | |
| | == LabelMapVolumeNode == | | == LabelMapVolumeNode == |
| − | * TBD
| |
| − | * Added labelmap node support to subject hierarchy
| |
| − | * Improved CLI image widget type handling.
| |
| | | | |
| | == Slice viewers == | | == Slice viewers == |
| − | * Added control to adjust thickness for label map outlines [http://www.na-mic.org/Bug/view.php?id=2267 #2267]
| |
| | | | |
| | == DataProbe == | | == DataProbe == |
| − | * Added magnified slice view into DataProbe panel.
| |
| | | | |
| | == SliceViewAnnotations == | | == SliceViewAnnotations == |
| − | * Added orientation marker to slice view annotations.
| |
| − | * Improved integration of SliceViewAnnotations. [http://www.na-mic.org/Bug/view.php?id=3943 #3943]
| |
| | | | |
| | == OpenIGTLink == | | == OpenIGTLink == |
| − | * Improved support for POINT and POLYDATA messages. [http://www.na-mic.org/Bug/view.php?id=3875 #3875] [http://www.na-mic.org/Bug/view.php?id=3907 #3907]
| |
| | | | |
| − | == DoubleArrays ==
| |
| − | * Improved loading and saving of measurement files (.mcsv)
| |
| − |
| |
| − | == CastScalarVolume ==
| |
| − | * Improved CastScalarVolume module to support input of type unsigned char, int or long.
| |
| − |
| |
| − | == SceneView ==
| |
| − | * Improved management of data loaded between creation of SceneViews
| |
| − |
| |
| − | == Landmark ==
| |
| − | * Added support for landmark refinement
| |
| − | * Improved LandmarkRegistration plugin loader displaying message if plugin failed to be loaded.
| |
| − |
| |
| − | == MultiVolumeImporter ==
| |
| − | * Fixed loading of MGH variable FA multivolume.
| |
| − |
| |
| − | == FiberBundleLabelSelect ==
| |
| − | * Fixed ADD operation for ‘Label to include’. [http://www.na-mic.org/Bug/view.php?id=4025 #4025]
| |
| − | * Added support for both VTK and VTP input files.
| |
| − |
| |
| − | == DTI support ==
| |
| − | * Resolved multithread issue that caused errors with multiple b values. [http://www.na-mic.org/Bug/view.php?id=3977 #3977]
| |
| − | * Fixed parallel/perpendicular diffusivity on glyphs, interactive seeding line display. [http://www.na-mic.org/Bug/view.php?id=3857 #3857]
| |
| − | * Fixed displaying of Tensor properties and their ranges in Tractography display module for lines/tubes/glyphs [http://www.na-mic.org/Bug/view.php?id=3857 #3857]
| |
| − | * Added logic for propagating color and opacity to fiber hierarchies. [http://www.na-mic.org/Bug/view.php?id=3925 #3925]
| |
| − | * Added support for Fiber Hierarchies in the Models Module. [http://www.na-mic.org/Bug/view.php?id=3678 #3678]
| |
| − |
| |
| − | == VolumeRendering ==
| |
| − | * Fixed volume rendering not visible when non-volume-rendered volume is frequently changing in the scene.
| |
| − | * Ensure volume rendering is updated when volume geometry is modified. [http://www.na-mic.org/Bug/view.php?id=4023 #4023]
| |
| − |
| |
| − | == FiberTractMeasurements ==
| |
| − | * TBD
| |
| − | * Added CLI module FiberTractMeasurements allowing to XXXX supporting cluster aggregation.
| |
| | | | |
| | = For Developers = | | = For Developers = |
| Line 301: |
Line 132: |
| | == Modules and Extensions == | | == Modules and Extensions == |
| | | | |
| | + | <!-- |
| | * ExtensionWizard | | * ExtensionWizard |
| − | ** Added "Enable developer mode" option to ExtensionWizard
| |
| | | | |
| | * SelfTests | | * SelfTests |
| Line 344: |
Line 175: |
| | ** Facilitate re-use of Editor python components in extension (LabelStructureListWidget in 043f398) | | ** Facilitate re-use of Editor python components in extension (LabelStructureListWidget in 043f398) |
| | ** Updated EditUtil API adding function SetUseLabelOutline() to explicitly set label outline state on all Slice nodes | | ** Updated EditUtil API adding function SetUseLabelOutline() to explicitly set label outline state on all Slice nodes |
| | + | --> |
| | | | |
| | == Slicer Core == | | == Slicer Core == |
| | | | |
| | + | <!-- |
| | * IDE integration | | * IDE integration |
| | ** Improve build targets organization in IDE that support folders. | | ** Improve build targets organization in IDE that support folders. |
| Line 394: |
Line 227: |
| | * DICOM | | * DICOM |
| | ** [https://github.com/Slicer/Slicer/pull/359 upgraded DCMTK to the latest snapshot DCMTK-3.6.1_20150924] | | ** [https://github.com/Slicer/Slicer/pull/359 upgraded DCMTK to the latest snapshot DCMTK-3.6.1_20150924] |
| − | | + | --> |
| | | | |
| | <gallery caption="Improved Toolkits" widths="350px" heights="250px" perrow="3"> | | <gallery caption="Improved Toolkits" widths="350px" heights="250px" perrow="3"> |
Summary
The community of Slicer developers is proud to announce the release of Slicer Nightly.
- Slicer Nightly introduces
- An improved App Store, known as the Extension Manager, for adding plug-ins to Slicer. More than 80 plug-ins and packages of plug-ins are currently available.
- Close to 150 feature improvements and bug fixes have resulted in improved performance and stability.
- Improvements to many modules.
- Click here to download Slicer Nightly for different platforms and find pointers to the source code, mailing lists and the bug tracker.
- Please note that Slicer continues to be a research package and is not intended for clinical use. Testing of functionality is an ongoing activity with high priority, however, some features of Slicer are not fully tested.
- The Slicer Training page provides a series of tutorials and data sets for training in the use of Slicer.
slicer.org is the portal to the application, training materials, and the development community.
What is 3D Slicer
3D Slicer is:
- A software platform for the analysis (including registration and interactive segmentation) and visualization (including volume rendering) of medical images and for research in image guided therapy.
- A free, open source software available on multiple operating systems: Linux, MacOSX and Windows
- Extensible, with powerful plug-in capabilities for adding algorithms and applications.
Features include:
- Multi organ: from head to toe.
- Support for multi-modality imaging including, MRI, CT, US, nuclear medicine, and microscopy.
- Bidirectional interface for devices.
There is no restriction on use, but Slicer is not approved for clinical use and intended for research. Permissions and compliance with applicable rules are the responsibility of the user. For details on the license see here
Citing Slicer
To acknowledge 3D Slicer as a platform, please cite the Slicer web site (http://www.slicer.org) and the following publication:
Fedorov A., Beichel R., Kalpathy-Cramer J., Finet J., Fillion-Robin J-C., Pujol S., Bauer C., Jennings D., Fennessy F., Sonka M., Buatti J., Aylward S.R., Miller J.V., Pieper S., Kikinis R. 3D Slicer as an Image Computing Platform for the Quantitative Imaging Network. Magnetic Resonance Imaging. 2012 Nov;30(9):1323-41. PMID: 22770690.
Slicer Nightly Highlights
- New and Improved Modules
Improved
Transforms module with support for non-linear transforms, visualization of transforms in 2D and 3D, detailed transform properties view -
click here for demo video.
- Added support for copy/paste of linear transforms
- Support saving of any transform to a grid transform
- Improved composite transform support
- Added support for thin plate spline transforms
- Addressed display issue for slice intersection display of non-linearly transformed model
The user interface of the
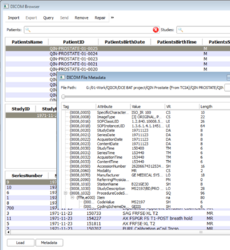
DICOM module has been improved for a better usability.
- Improved DICOM browser adding a right click menu to allow deleting or exporting to disk at the patients/studies/series levels.
- Added support for DICOM private dictionary.
- Improved support for loading of DICOM referenced datasets.
- Read and apply window/level found in DICOM volumes.
- Added support for Philips DWI datasets
- Fixed loading of DICOM image from files containing special characters.
Improved
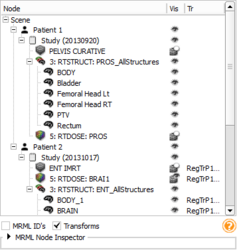
Subject hierarchy module for organizing and manipulating data loaded in Slicer.
- Highlight nodes in subject hierarchy referenced by DICOM
- Added Transforms subject hierarchy plugin
- Added new DICOM export feature and scalar volume plugin. #3163
- Added generic folder plugin to subject hierarchy
- Subject hierarchy nodes automatically created for supported types
- Added auto-delete subject hierarchy children to Application Settings
- Added auto-create subject hierarchy option to Application Settings
- Subject hierarchy tree headers show icons instead of text
- Fixed subject hierarchy nodes creation when importing old scene #3902
- Fixed volume visibility controls. #3893
Slicer Training
The Slicer Training page provides a series of updated tutorials and data sets for training in the use of Slicer Nightly.
The first 3D Slicer training events using Slicer 4.5 will be organized at Massachusetts General Hospital (MGH), Boston, MA, Brown University, Providence, RI and RSNA 2015, Chicago, Il.
Slicer Extensions
- New Extensions
AnglePlanes This Module is used to calculate the angle between two planes by using the normals NEW
Improved Extensions in Slicer 4.5
Extensions removed from Slicer 4.5
Extensions renamed
Other Improvements, Additions & Documentation
Optimization
Transforms
DICOM
Data processing
CLI
Usability
SubjectHierarchy
Python scripting
Editor
Markups
LabelMapVolumeNode
Slice viewers
DataProbe
SliceViewAnnotations
OpenIGTLink
For Developers
Modules and Extensions
Slicer Core
- Improved Toolkits
Moved from CTKAppLauncher v0.1.11 to v0.1.14 (43 commits)
Moved from ITK v4.4.1 to v4.6.0 (1089 commits)
Moved from Qt 4.7.4 to Qt 4.8.6
Moved from VTK v5.10.1 to VTK v6.2.0 (5490 commits)
Looking at the Code Changes
From a git checkout you can easily see the all the commits since the time of the 4.4.0 release:
git log v4.4.0..HEAD
To see a summary of your own commits, you could use something like:
git log v4.4.0..HEAD --oneline --author=me
see the git log man page for more options.
Commit stats and full changelog
Related Projects