Difference between revisions of "Documentation/4.4/gif tutorial v3 1"
From Slicer Wiki
| Line 2: | Line 2: | ||
{| class="wikitable" style="border-spacing: 2px; border: 1px solid darkgray;" | {| class="wikitable" style="border-spacing: 2px; border: 1px solid darkgray;" | ||
| − | | | + | ! scope="col" | Table of Contents |
|- | |- | ||
| − | |[[Documentation/4.4/gif tutorial v3 2|Segmentation]] | + | |[[Documentation/4.4/gif tutorial v3 2|2. Segmentation]] |
|- | |- | ||
| − | |[[Documentation/4.4/gif tutorial v3 3|Tractography exploration of peritumoral white matter fibers]] | + | |[[Documentation/4.4/gif tutorial v3 3|3. Tractography exploration of peritumoral white matter fibers]] |
|- | |- | ||
| − | |[[Documentation/4.4/gif tutorial v3 4|Tractography exploration of the ipsilateral and contralateral side]] | + | |[[Documentation/4.4/gif tutorial v3 4|4. Tractography exploration of the ipsilateral and contralateral side]] |
|} | |} | ||
Latest revision as of 20:26, 20 July 2015
Home < Documentation < 4.4 < gif tutorial v3 1|
| Table of Contents |
|---|
| 2. Segmentation |
| 3. Tractography exploration of peritumoral white matter fibers |
| 4. Tractography exploration of the ipsilateral and contralateral side |
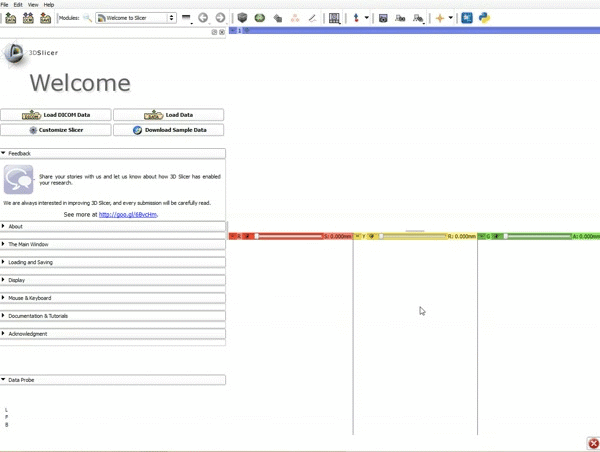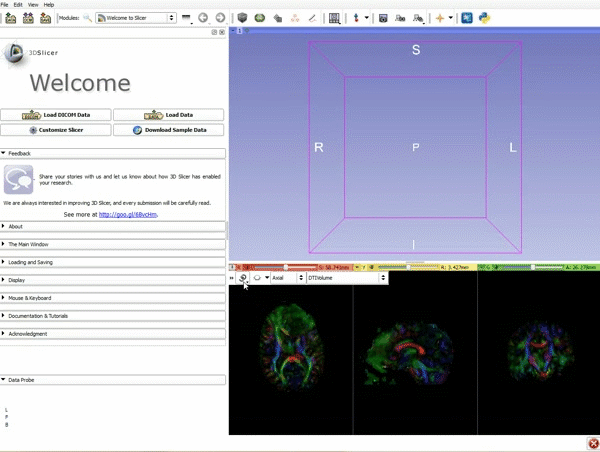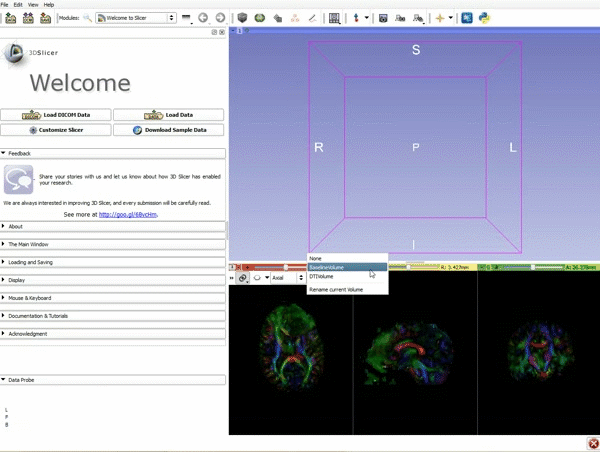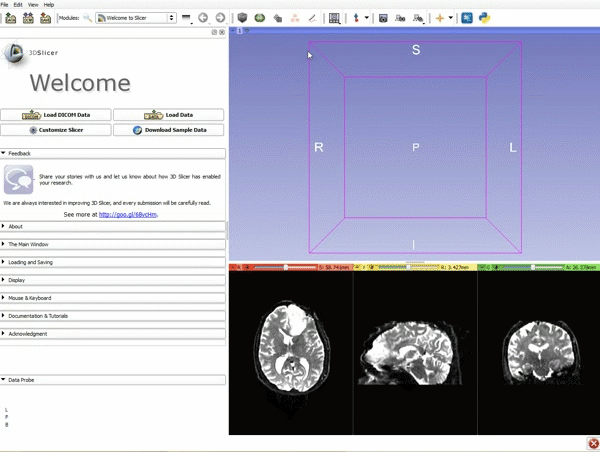
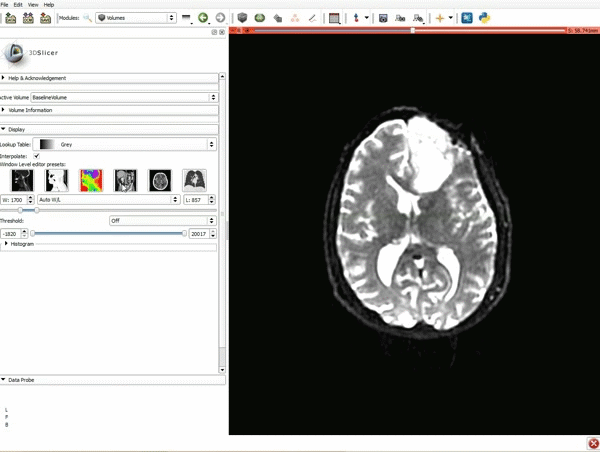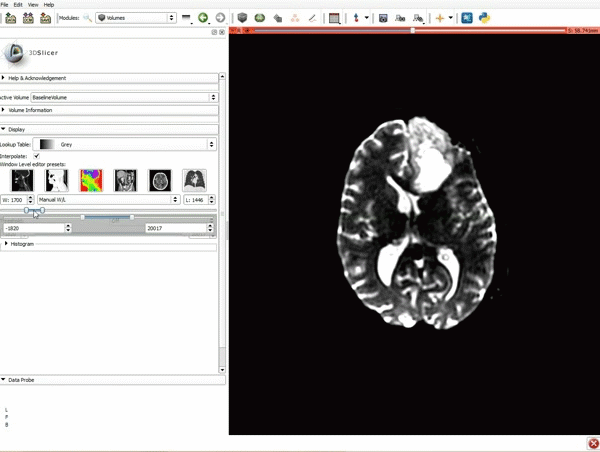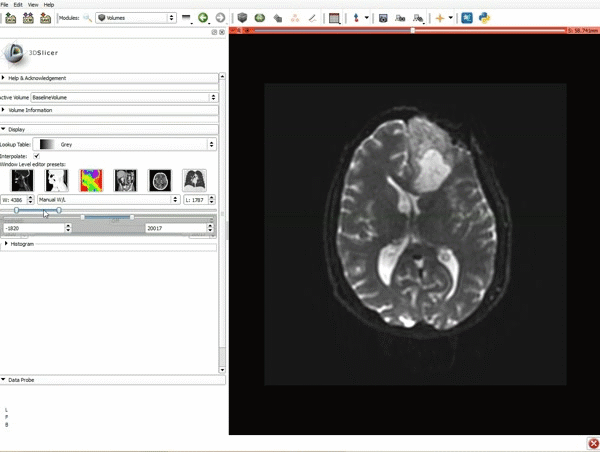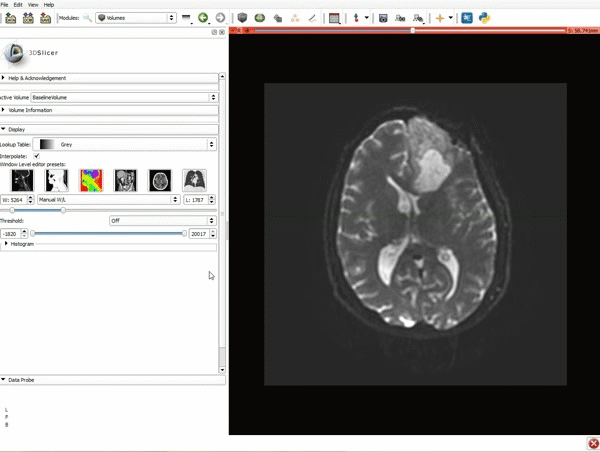
Loading DTI and Baseline Data
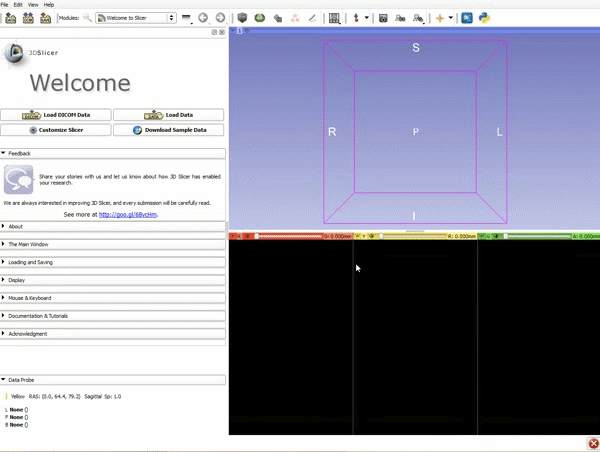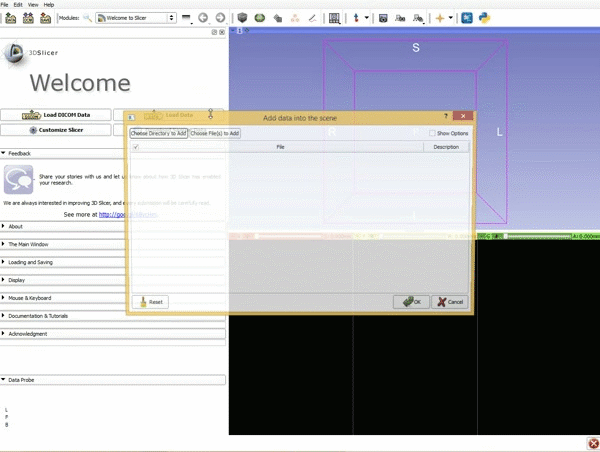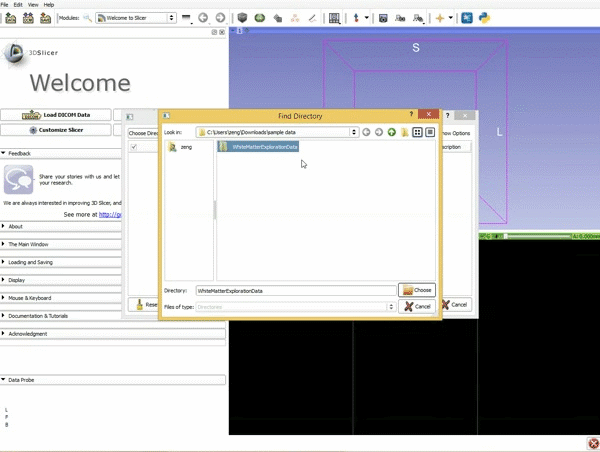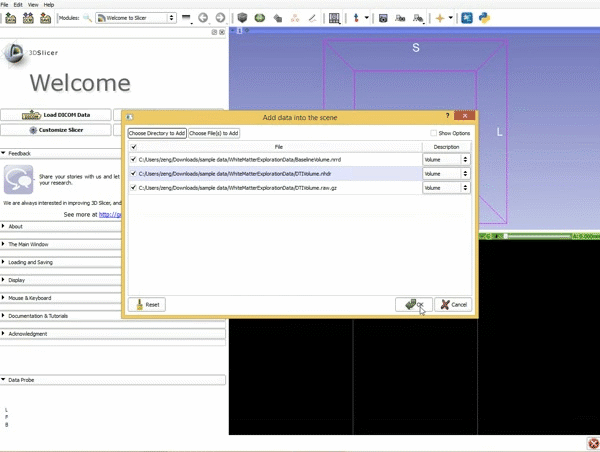
| 1. Load the sample data by selecting "Load Data", then "Choose Directory to Add" and finally going to wherever it is that you downloaded the sample data and selecting the folder WhiteMatterExplorationData | 2. Click on the pin icon to display the slice menu,then click on the link icon to link the 3 anatomical viewers. Then change the background so it is set to BaselineVolume. |
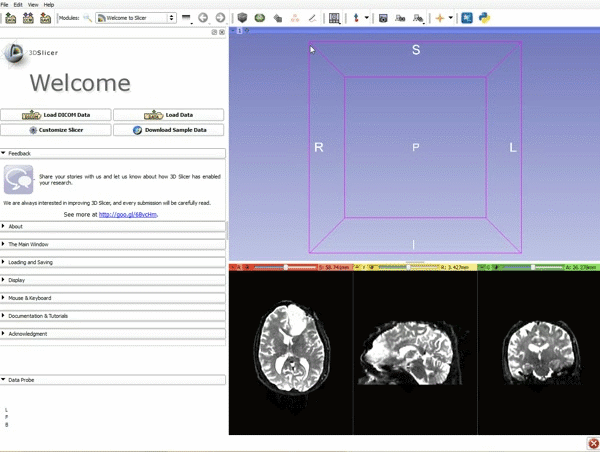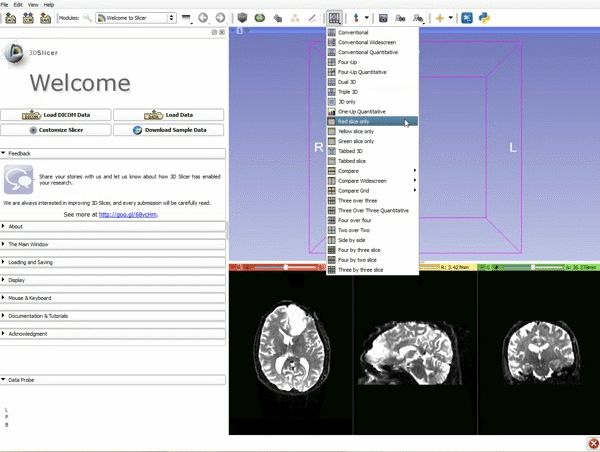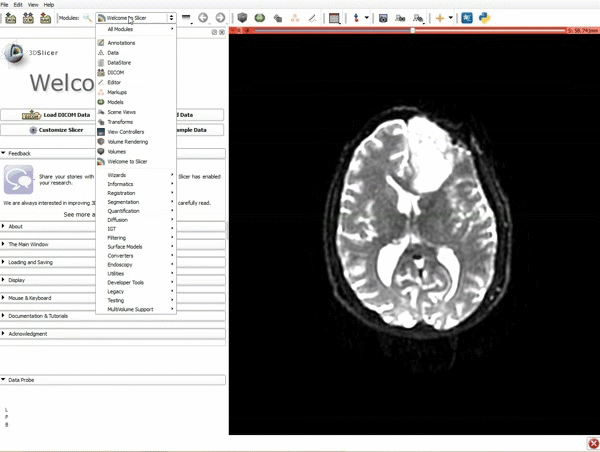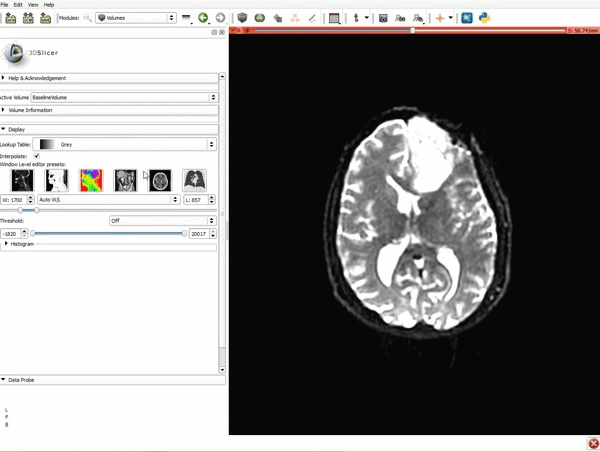
| 3. a) Click on the Layout Menu and select Red Slice Only. b) Select the Volumes Module from the Modules menu | 4. Manually adjust the Window Level editor presets with the Volume module menu |