Difference between revisions of "Documentation/Nightly/Modules/PkModeling"
| Line 10: | Line 10: | ||
Extension: [[Documentation/{{documentation/version}}/Extensions/PkModeling|PkModeling]]<br> | Extension: [[Documentation/{{documentation/version}}/Extensions/PkModeling|PkModeling]]<br> | ||
Acknowledgments: | Acknowledgments: | ||
| − | This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, and by National Cancer Institute as part of the Quantitative Imaging Network initiative (U01CA151261).<br> | + | This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, and by National Cancer Institute as part of the Quantitative Imaging Network initiative (U01CA151261) and QIICR (U24CA180918).<br> |
Implementation of the pharmacokinetics modeling was contributed by Yingxuan Zhu and Jim Miller from GE Research.<br> | Implementation of the pharmacokinetics modeling was contributed by Yingxuan Zhu and Jim Miller from GE Research.<br> | ||
Author: Yingxuan Zhu (while at GE), Andrey Fedorov (SPL), Jim Miller ({{collaborator|name|ge}}), John Evans (MGH)<br> | Author: Yingxuan Zhu (while at GE), Andrey Fedorov (SPL), Jim Miller ({{collaborator|name|ge}}), John Evans (MGH)<br> | ||
Revision as of 03:08, 29 May 2014
Home < Documentation < Nightly < Modules < PkModeling
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: PkModeling | |||||
|
Module Description
|
PkModeling (Pharmacokinetics Modeling) calculates quantitative parameters from Dynamic Contrast Enhanced DCE-MRI images. This module performs two operations:
|
Use Cases
Tutorials
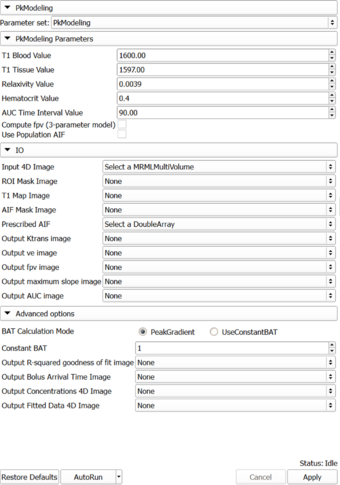
Panels and their use
|
The following acquisition parameters should be available in the NRRD header of the input data (if you are analyzing a DICOM time series, they will typically be extracted from the DICOM data):
- TR Value: Repetition time (milliseconds)
- TE Value: Echo time (milliseconds)
- FA Value: Flip angle (degrees)
- Timestamps for the dynamic series (in milliseconds)
Here is an example how this information is represented in the NRRD header:
MultiVolume.DICOM.EchoTime:=2.93 MultiVolume.DICOM.FlipAngle:=10 MultiVolume.DICOM.RepetitionTime:=6.13 MultiVolume.FrameIdentifyingDICOMTagName:=AcquisitionTime MultiVolume.FrameIdentifyingDICOMTagUnits:=ms
Similar Modules
References
- [1] Knopp MV, Giesel FL, Marcos H et al: Dynamic contrast-enhanced magnetic resonance imaging in oncology. Top Magn Reson Imaging, 2001; 12:301-308.
- [2] Rijpkema M, Kaanders JHAM, Joosten FBM et al: Method for quantitative mapping of dynamic MRI contrast agent uptake in human tumors. J Magn Reson Imaging 2001; 14:457-463.
- [3] de Bazelaire, C.M., et al., MR imaging relaxation times of abdominal and pelvic tissues measured in vivo at 3.0 T: preliminary results. Radiology, 2004. 230(3): p. 652-9.
- [4] Pintaske J, Martirosian P, Graf H, Erb G, Lodemann K-P, Claussen CD, Schick F. Relaxivity of Gadopentetate Dimeglumine (Magnevist), Gadobutrol (Gadovist), and Gadobenate Dimeglumine (MultiHance) in human blood plasma at 0.2, 1.5, and 3 Tesla. Investigative radiology. 2006 March;41(3):213–21.
- [5] Parker GJ, Roberts C, Macdonald A, Buonaccorsi GA, Cheung S, Buckley DL, Jackson A, Watson Y, Davies K, Jayson GC. Experimentally-derived functional form for a population-averaged high-temporal-resolution arterial input function for dynamic contrast-enhanced MRI. Magnetic Resonance in Medicine, 2006 Nov; 56(5):993-1000.
Information for Developers
| Section under construction. |