Difference between revisions of "Documentation/Nightly/Modules/PkModeling"
From Slicer Wiki
| Line 56: | Line 56: | ||
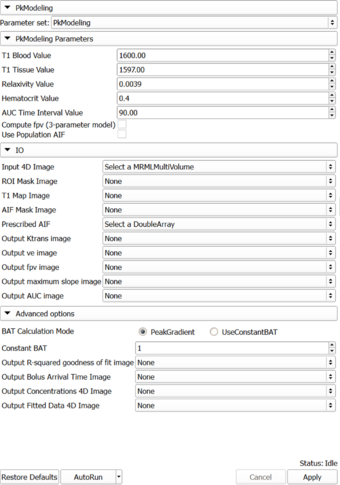
*** T1 Blood Value | *** T1 Blood Value | ||
*** T1 Tissue Value (default value is the published value for prostate tissue estimated in healthy individuals (see Ref. de Bazelaire et al.) | *** T1 Tissue Value (default value is the published value for prostate tissue estimated in healthy individuals (see Ref. de Bazelaire et al.) | ||
| − | *** Relaxivity Value | + | *** Relaxivity Value (contrast agent specific, default value corresponds to Gd DPTA) |
*** Hematocrit Value. Volume percentage of red blood cells in blood. | *** Hematocrit Value. Volume percentage of red blood cells in blood. | ||
*** AUC Time Interval Value: Time interval for AUC calculation | *** AUC Time Interval Value: Time interval for AUC calculation | ||
Revision as of 16:43, 23 May 2013
Home < Documentation < Nightly < Modules < PkModelingIntroduction and Acknowledgements
|
Extension: PkModeling | |||||
|
Module Description
|
PkModeling (Pharmacokinetics Modeling) calculates quantitative parameters from Dynamic Contrast Enhanced DCE-MRI images. This module performs two operations:
|
Use Cases
Tutorials
Panels and their use
|
Similar Modules
References
- [1] Knopp MV, Giesel FL, Marcos H et al: Dynamic contrast-enhanced magnetic resonance imaging in oncology. Top Magn Reson Imaging, 2001; 12:301-308.
- [2] Rijpkema M, Kaanders JHAM, Joosten FBM et al: Method for quantitative mapping of dynamic MRI contrast agent uptake in human tumors. J Magn Reson Imaging 2001; 14:457-463.
- [3] de Bazelaire, C.M., et al., MR imaging relaxation times of abdominal and pelvic tissues measured in vivo at 3.0 T: preliminary results. Radiology, 2004. 230(3): p. 652-9.
Information for Developers
| Section under construction. |