Difference between revisions of "EMSegmenter-Tasks:Human-Eye"
Belhachemi (talk | contribs) (→Result) |
(→Atlas) |
||
| Line 26: | Line 26: | ||
=Atlas= | =Atlas= | ||
| − | Atlas was generated based on 5 scans and corresponding segmentations provided by Raphaël Olszewski, l'Université catholique de Louvain à Louvain-La-Neuve, Belgique [link]. We registered the scans to a preselected template via Warfield et al. 2001. <BR> | + | Atlas was generated based on 5 scans and corresponding segmentations provided by Raphaël Olszewski, l'Université catholique de Louvain à Louvain-La-Neuve, Belgique [link]. We registered the scans to a preselected template via (Warfield et al. 2001.) <BR> |
Image Dimension = 256 x 256 x 100 <br> | Image Dimension = 256 x 256 x 100 <br> | ||
Image Spacing = 1 x 1 x 1 <BR> | Image Spacing = 1 x 1 x 1 <BR> | ||
| Line 33: | Line 33: | ||
Image:EMS HumanEyeAtlasLabels.png | Image:EMS HumanEyeAtlasLabels.png | ||
</gallery> | </gallery> | ||
| − | |||
=Result= | =Result= | ||
Revision as of 14:08, 27 April 2011
Home < EMSegmenter-Tasks:Human-EyeReturn to EMSegmenter Task Overview Page
Contents
Description
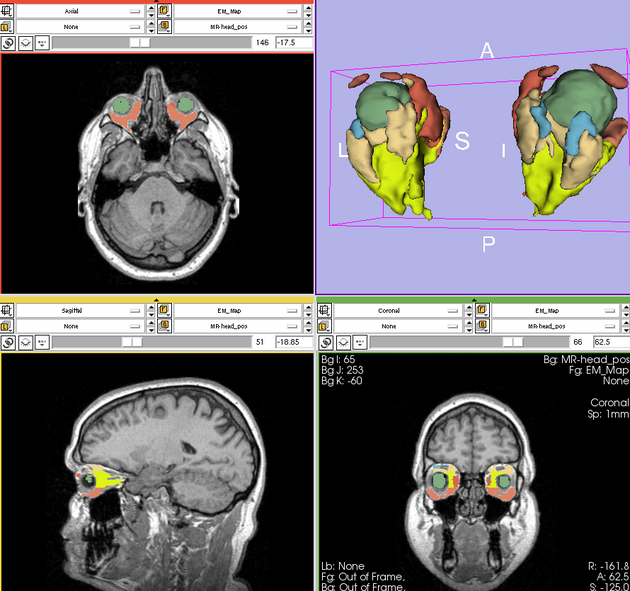
Single channel automatic segmentation of t1w-MRI brain scans into the major tissue classes (gray matter, white matter, csf). The task can only be applied to t1w brain scan showing parts of the skull and neck. The pipeline consist of the following steps:
- Step 1: Perform image inhomogeneity correction of the MRI scan via N4ITKBiasFieldCorrection (Tustison et al 2010)
- Step 2: Register the atlas to the MRI scan via BRAINSFit (Johnson et al 2007)
- Step 3: Compute the intensity distributions for each structure
Compute intensity distribution (mean and variance) for each label by automatically sampling from the MR scan. The sampling for a specific label is constrained to the region that consists of voxels with high probability (top 95%) of being assigned to the label according to the aligned atlas.
- Step 4: Automatically segment the MRI scan into the structures of interest using EM Algorithm (Pohl et al 2007)
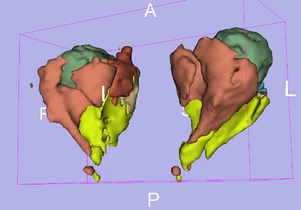
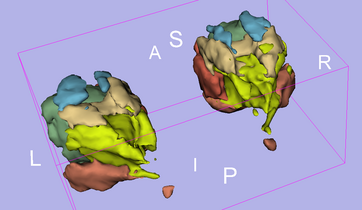
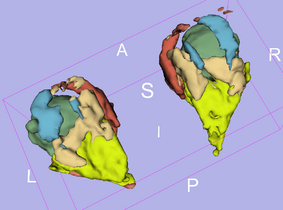
Anatomical Tree
labels/colors need to be updated
- root
- 1 (dark blue) : not used
- 2 (pink) : fat tissue
- 3 (white) : not used
- 4 (blue) : inferior extraocular muscle
- 5 (red) : medial extraocular muscle
- 6 (green) : superior extraocular muscle
- 7 (azure blue) : superior oblique extraocular muscle
- 8 (olive green) : lateral extraocular muscle
- 9 (violet) : inferior extraocular muscle
- 10 (olive green) : non used
Atlas
Atlas was generated based on 5 scans and corresponding segmentations provided by Raphaël Olszewski, l'Université catholique de Louvain à Louvain-La-Neuve, Belgique [link]. We registered the scans to a preselected template via (Warfield et al. 2001.)
Image Dimension = 256 x 256 x 100
Image Spacing = 1 x 1 x 1
Result
Collaborators
Raphaël Olszewski, l'Université catholique de Louvain à Louvain-La-Neuve, Belgique
Acknowledgment
The construction of the pipeline was supported by funding from NIH NCRR 2P41RR013218 Supplement.
Citations
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC N4ITK: Improved N3 Bias Correction, IEEE Trans Med Imag, 2010
- Pohl K, Bouix S, Nakamura M, Rohlfing T, McCarley R, Kikinis R, Grimson W, Shenton M, Wells W. A Hierarchical Algorithm for MR Brain Image Parcellation. IEEE Transactions on Medical Imaging. 2007 Sept;26(9):1201-1212.
- S. Warfield, J. Rexilius, P. Huppi, T. Inder, E. Miller, W. Wells, G. Zientara, F. Jolesz, and R. Kikinis, “A binary entropy measure to assess nonrigid registration algorithms,” in MICCAI, LNCS, pp. 266–274, Springer, October 2001.
- Johnson H.J., Harris G., Williams K. BRAINSFit: Mutual Information Registrations of Whole-Brain 3D Images, Using the Insight Toolkit, The Insight Journal, July 2007