Difference between revisions of "Slicer 3.6:Training"
From Slicer Wiki
(→Slicer 3.6 Tutorials: Fix broken link) |
Belhachemi (talk | contribs) (Added EMSegmenter Tutorials) |
||
| Line 67: | Line 67: | ||
| style="background:#FFFFCC; color:black" valign="top" | [http://wiki-na-mic.org/Wiki/images/7/73/PETCTFusion-Tutorial-Data.zip '''PET/CT Data ''' (zip archive)] | | style="background:#FFFFCC; color:black" valign="top" | [http://wiki-na-mic.org/Wiki/images/7/73/PETCTFusion-Tutorial-Data.zip '''PET/CT Data ''' (zip archive)] | ||
| style="background:#FFFFCC; color:black" align="center"| [[Image:PETCTFusionBig.png|250px]] | | style="background:#FFFFCC; color:black" align="center"| [[Image:PETCTFusionBig.png|250px]] | ||
| + | |- | ||
| + | | style="background:#FFFFCC; color:blue; font-size:110%" align="center"| <span id="1.1"></span> '''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top"| [http://www.slicer.org/slicerWiki/images/d/d9/EMSegmenterTutorialSimpleMode-2010-Nov.pdf '''EMSegmenter Simple Mode'''] <br><br> This tutorial takes the trainee through the segmentation of a MRI Human Brain without to adjust any parameters. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" | [http://www.slicer.org/slicerWiki/images/c/cd/MRIHumanBrain_T1_aligned.nrrd '''Human Brain T1 Data'''] | ||
| + | | style="background:#FFFFCC; color:black" align="center"| [[Image:EMSegmenter-Simple-Mode.png|250px]] | ||
| + | |- | ||
| + | | style="background:#FFFFCC; color:blue; font-size:110%" align="center"| <span id="1.1"></span> '''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top"| [http://www.slicer.org/slicerWiki/images/b/b8/EMSegmenterTutorialAdvancedMode-2010-Nov.pdf '''EMSegmenter Advanced Mode'''] <br><br> This tutorial takes the trainee through the segmentation of a MRI Human Brain. The trainee will learn how to setup the EMSegmenter, this includes the creation of a task, the creation of an anatomical tree and adjusting the weights for the EM algorithm. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" | [http://www.slicer.org/slicerWiki/images/c/cd/MRIHumanBrain_T1_aligned.nrrd '''Human Brain T1 Data'''] | ||
| + | | style="background:#FFFFCC; color:black" align="center"| [[Image:EMSegmenter-Advanced-Mode.png|250px]] | ||
|} | |} | ||
Revision as of 17:57, 24 November 2010
Home < Slicer 3.6:TrainingContents
Slicer 3.6 Tutorials
- The following table contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment (version 3.6 release) to accomplish certain tasks.
- For tutorials for other versions of Slicer, please visit the Slicer training portal.
- For questions related to the Slicer3 Compendium, please send an e-mail to Sonia Pujol, Ph.D
| Category | Tutorial | Sample Data | Image |
| Core | Slicer3Minute Tutorial-PDF Slicer3Minute Tutorial-PPT The Slicer3Minute tutorial is an introduction to the advanced 3D visualization capabilities of Slicer3.6. Audience: First time users. |
Slicer3Minute Data The Slicer3Minute dataset contains an MR scan of the brain and 3D reconstructions of the anatomy |
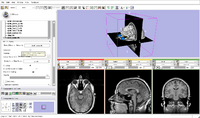
|
| Core | Slicer3Visualization Tutorial-PDF Slicer3Visualization Tutorial-PPT The Slicer3Visualization tutorial guides through 3D data loading and visualization in Slicer3.6. Audience: All beginners including clinicians, scientists, engineers and programmers. |
Slicer3Visualization Data The Slicer3VisualizationDataset contains two MR scans of the brain, a pre-computed labelmap and 3D reconstructions of the anatomy. |
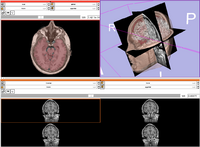
|
| Core | Programming in Slicer3 Tutorial-PDF Programming in Slicer3 Tutorial-PPT The Programming in Slicer3 tutorial is an introduction to the integration of C++ stand-alone programs outside of the Slicer3 source tree. Audience: Programmers and Engineers. |
HelloWorld Plugin The HelloWorld tutorial dataset contains an MR scan of the brain and pre-computed xml and C++ files for integrating the Hello Python plug-in to Slicer3. |
|
| Core | Hello Python Tutorial The Hello Python tutorial is an introduction to the integration of Python stand-alone programs outside of the Slicer3 source tree. Audience: Programmers and Engineers. |
HelloPython Plugin The HelloPython tutorial dataset contains an MR scan of the brain and pre-computed xml and Python code for integrating the Hello Python plug-in to Slicer3. |
|
| Core | Interactive Editor-PDF Interactive Editor-PPT Shows how to use the interactive editing tools in Slicer. Audience: All users and developers. |
Editor Data This dataset contains a MR dataset of the brain. |
|
| Core | Manual Registration-PDF Manual Registration-PPT Shows how to manually/interactively align two images in Slicer3.6 Audience: First time & early users. |
Manual Registration Data This dataset contains two brain MRI of a single subject, obtained in different orientations. |

|
| Specialized | Diffusion MRI tutorial-PDF Diffusion MRI tutorial-PPT This tutorial guides you through the process of loading diffusion MR data, estimating diffusion tensors, and performing tractography of white matter bundles. Audience: All users and developers. |
Diffusion Data | 
|
| Specialized | Change Tracker Tutorial-PDF Change Tracker Tutorial-PPT This tutorial describes the use of ChangeTracker module to detect changes in tumor volume from two MRI scans. Audience: All users interested in longitudinal analysis of pathology. |
Training Data download is integrated with the ChangeTracker module (see Tutorial) | |
| Specialized | FreeSurfer Course The FreeSurfer dataset contains an MR scan of the brain and pre-computed FreeSurfer segmentation and cortical surface reconstructions.
|
FreeSurfer tutorial data | 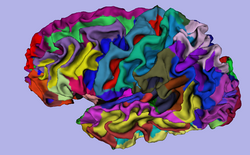
|
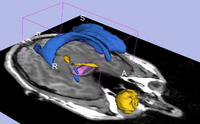
| Specialized | Neurosurgical Planning Tutorial This tutorial takes the trainee through a complete workup for neurosurgical patient-specific mapping. Also see this tutorial for information on how to use Slicer's affine registration, simple region growing, model maker and tractography modules. Audience: All users interested in image-guided therapy. |
Neurosurgical Planning Data | 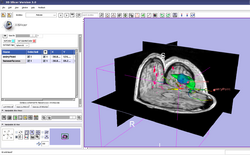
|
| Specialized | PET/CT SUV Tutorial This tutorial takes the trainee through the computation of SUV body weight on a baseline and followup study. |
PET/CT Data (zip archive) | 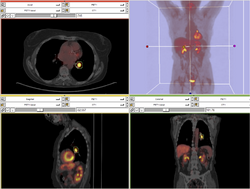
|
| Specialized | EMSegmenter Simple Mode This tutorial takes the trainee through the segmentation of a MRI Human Brain without to adjust any parameters. |
Human Brain T1 Data | 
|
| Specialized | EMSegmenter Advanced Mode This tutorial takes the trainee through the segmentation of a MRI Human Brain. The trainee will learn how to setup the EMSegmenter, this includes the creation of a task, the creation of an anatomical tree and adjusting the weights for the EM algorithm. |
Human Brain T1 Data | 
|
Summer 2010 Tutorial Contest Entries (under construction)
The following tutorials were part of the Summer 2010 Slicer Tutorial Contest.
| Category | Tutorial | Sample Data | Image |
| Specialized | Meshing Workflow tutorial Audience: All users and developers. (Note: Mac and Linux only.) |
Meshing Workflow Data |
|
| Specialized | Fiducials tutorial Audience: All users and developers. |
Fiducials Data |
|
| Specialized | Robust Statistic Segmenter Audience: All users and developers. |
Robust Statistic Segmenter Data |
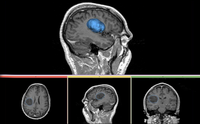
|
| Specialized | Longitudinal lesion comparison Audience: All users and developers. (Note: Mac and Linux only.) |
Longitudinal Lesion Comparison Data | 
|
| Specialized | Robot-Assisted MRI-Guided Prostate Biopsy Audience: All users and developers. |
Robot-Assisted MRI-Guided Prostate Biopsy | 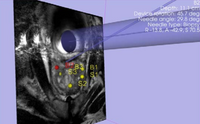
|
| Specialized | Atlas Label Fusion & Surface Registration Audience: All users and developers. |
Atlas Label Fusion & Surface Registration | 
|
| Specialized | Stochastic Tractography Audience: All users and developers. (Note: Mac and Linux only.) |
Stochastic Tractography | 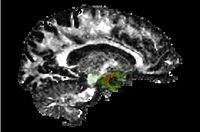
|
| Specialized | PERK Station Module Audience: All users and developers. (Note: Windows only.) |
PERK Station Module | 
|
Software Installation
- The Slicer download page contains information on how to obtain a compiled version of Slicer for a variety of platforms and where to find the source code for Slicer 3.
Software Documentation
- For the Slicer 3.6 manual pages please click here. These pages are the reference manual for Slicer 3.6 and briefly explain the functionality found in panels and modules.