Difference between revisions of "Modules:N4ITKBiasFieldCorrection-Documentation-3.6"
(Created page with 'Return to Slicer 3.6 Documentation Gallery of New Features __NOTOC__ ===N4ITK Bias field correction=== {| |[[Image…') |
|||
| Line 25: | Line 25: | ||
===Module Description=== | ===Module Description=== | ||
| − | This module is a CLI wrapper around the N4 bias field correction algorithm, which was presented by Nick Tustison in [http://hdl.handle.net/10380/3053 this Insight Journal publication]. | + | This module is a CLI wrapper around the N4 bias field correction algorithm, which was presented by Nick Tustison in [http://hdl.handle.net/10380/3053 this Insight Journal publication]. This module can be used to remove field inhomogeneity artifact from the image. |
== Usage == | == Usage == | ||
| + | With the basic usage scenario, you only need to specify the input and output images. You can improve the performance of this module by specifying the binary mask for the region of interest in your input image. If this mask is not specified, the module will use Otsu thresholding algorithm to estimate the mask automatically. | ||
| + | In case you are not happy with the result of processing, you may need to experiment with the parameters described below. If you are still not satisfied with the result, you will need to study the relevant papers (see References section at the bottom of this page) and/or contact the Slicer and ITK user lists. | ||
===Use Cases, Examples=== | ===Use Cases, Examples=== | ||
| Line 35: | Line 37: | ||
This module is especially appropriate for these use cases: | This module is especially appropriate for these use cases: | ||
| − | * | + | * You observe smooth variation of the intensity over the tissue that should have intensity close to uniform |
| − | * | + | * Your attempts to segment or register your data are not successful, and you are not sure what to do next |
Examples of the module in use: | Examples of the module in use: | ||
| Line 45: | Line 47: | ||
===Tutorials=== | ===Tutorials=== | ||
| − | + | There are no tutorials available at this time. | |
| − | * | + | However, you can use the images that we use to test the functionality of this module, and the output it generates with the default values of the parameters to experiment with the parameters: |
| − | ** Data | + | * [http://viewvc.slicer.org/viewcvs.cgi/*checkout*/trunk/Testing/Data/Input/he3volume.nii.gz?rev=12975 input image] |
| + | * [http://viewvc.slicer.org/viewcvs.cgi/*checkout*/trunk/Testing/Data/Input/he3mask.nii.gz?rev=12975 input image mask] | ||
| + | * [http://viewvc.slicer.org/viewcvs.cgi/*checkout*/trunk/Testing/Data/Baseline/CLI/he3corrected.nii.gz?rev=12975 output image] | ||
| + | * [http://viewvc.slicer.org/viewcvs.cgi/*checkout*/trunk/Testing/Data/Baseline/CLI/he3biasfield.nii.gz?rev=12975 recovered bias field] | ||
| + | |||
| + | {| | ||
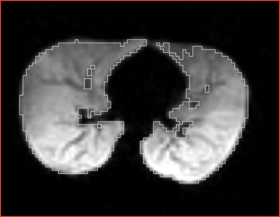
| + | |[[Image:n4_he_input.png|thumb|280px|Slice of the input test volume with bias field]] | ||
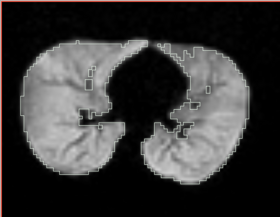
| + | |[[Image:n4_he_output.png|thumb|280px|Output image with bias removed]] | ||
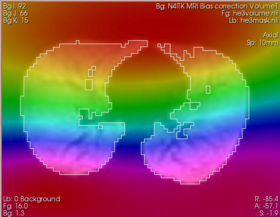
| + | |[[Image:n4_he_bias.png|thumb|280px|Recovered bias field (zoom in to see the actual value at the mouse pointer -- the field is multiplicative)]] | ||
| + | |} | ||
===Quick Tour of Features and Use=== | ===Quick Tour of Features and Use=== | ||
Revision as of 19:24, 28 April 2010
Home < Modules:N4ITKBiasFieldCorrection-Documentation-3.6Return to Slicer 3.6 Documentation
N4ITK Bias field correction
General Information
Module Type & Category
Type: CLI
Category: Experimental
Authors, Collaborators & Contact
- Author: Nick Tustison, UPenn (algorithm and ITK implementation)
- Author: Andriy Fedorov, BWH (Slicer integration)
- Contact: Andriy Fedorov, fedorov at bwh dot harvard dot edu
Module Description
This module is a CLI wrapper around the N4 bias field correction algorithm, which was presented by Nick Tustison in this Insight Journal publication. This module can be used to remove field inhomogeneity artifact from the image.
Usage
With the basic usage scenario, you only need to specify the input and output images. You can improve the performance of this module by specifying the binary mask for the region of interest in your input image. If this mask is not specified, the module will use Otsu thresholding algorithm to estimate the mask automatically.
In case you are not happy with the result of processing, you may need to experiment with the parameters described below. If you are still not satisfied with the result, you will need to study the relevant papers (see References section at the bottom of this page) and/or contact the Slicer and ITK user lists.
Use Cases, Examples
This module is especially appropriate for these use cases:
- You observe smooth variation of the intensity over the tissue that should have intensity close to uniform
- Your attempts to segment or register your data are not successful, and you are not sure what to do next
Examples of the module in use:
- Example 1
- Example 2
Tutorials
There are no tutorials available at this time.
However, you can use the images that we use to test the functionality of this module, and the output it generates with the default values of the parameters to experiment with the parameters:
Quick Tour of Features and Use
A list panels in the interface, their features, what they mean, and how to use them. For instance:
|
Development
Notes from the Developer(s)
Algorithms used, library classes depended upon, use cases, etc.
Dependencies
Other modules or packages that are required for this module's use.
Tests
On the Dashboard, these tests verify that the module is working on various platforms:
- MyModuleTest1 MyModuleTest1.cxx
- MyModuleTest2 MyModuleTest2.cxx
Known bugs
Links to known bugs in the Slicer3 bug tracker
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Links to the module's source code:
Source code:
Doxygen documentation:
More Information
Acknowledgment
Include funding and other support here.
References
Publications related to this module go here. Links to pdfs would be useful.