Difference between revisions of "Modules:ARCTIC-Documentation-3.6"
(→Tests) |
|||
| Line 51: | Line 51: | ||
{| | {| | ||
| | | | ||
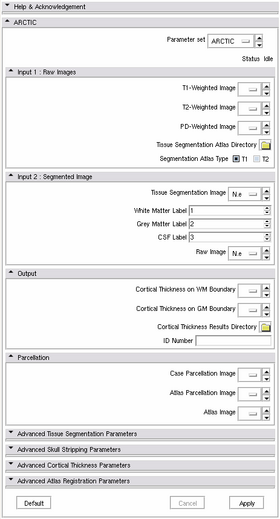
| + | [[Image:ARCTIC.png|thumb|280px|ARCTIC User Interface]] | ||
* '''Input 1 - Raw Images:''' set input images representing the different modalities of the same subject | * '''Input 1 - Raw Images:''' set input images representing the different modalities of the same subject | ||
** T1-weighted image: set input T1 image, if available | ** T1-weighted image: set input T1 image, if available | ||
| Line 80: | Line 81: | ||
*** Atlas parcellation image: set the atlas parcellation image | *** Atlas parcellation image: set the atlas parcellation image | ||
*** Atlas image: set the grayscale atlas image | *** Atlas image: set the grayscale atlas image | ||
| − | |||
| + | [[Image:ARCTIC_AdvancedParams.png|thumb|280px|User Interface]] | ||
* '''Advanced tissue segmentation parameters''' | * '''Advanced tissue segmentation parameters''' | ||
** Filter options: specifies smoothing parameters prior to the segmentation | ** Filter options: specifies smoothing parameters prior to the segmentation | ||
| Line 102: | Line 103: | ||
** Choose between different initialization methods : see registration pipeline discussion | ** Choose between different initialization methods : see registration pipeline discussion | ||
| − | |||
| − | |||
|} | |} | ||
Revision as of 21:31, 6 April 2010
Home < Modules:ARCTIC-Documentation-3.6Return to Slicer 3.6 Documentation
ARCTIC: Automatic Regional Cortical ThICkness
General Information
Module Type & Category
Type: CLI
Category: Workflow
Authors, Collaborators & Contact
- Author1: Cedric Mathieu, UNC-Chapel Hill
- Contributor1: Clement Vachet, UNC-Chapel Hill
- Contributor2: Martin Styner, UNC-Chapel Hill
- Contributor3: Heather Cody Hazlett, UNC-Chapel Hill
- Contact: Clement Vachet, cvachet[at]email[dot]unc[dot]edu
Module Description
ARCTIC (Automatic Regional Cortical ThICkness) is an end-to-end application developped at UNC-Chapel Hill allowing individual analysis of cortical thickness. This cross-platform tool can be run within Slicer3 as an external module, or directly as a command line.
Usage
Use Cases, Examples
This module is especially appropriate when one wants to perform individual regional cortical thickness analysis. ARCTIC allows efficient QC via precomputed 3D-Slicer scenes.
Tutorials
- Tutorials:
- Tutorial Dataset
- Wiki page on NITRC
Quick Tour of Features and Use
|
Development
Notes from the Developer(s)
Algorithms used, library classes depended upon, use cases, etc.
Dependencies
- Slicer3 modules:
- Publicly available atlases on MIDAS:
Tests
On the Dashboard, these tests verify that the module is working on various platforms:
- MyModuleTest1 MyModuleTest1.cxx
- MyModuleTest2 MyModuleTest2.cxx
Known bugs
Links to known bugs in the Slicer3 bug tracker
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
- Source code
- Download release source code via svn:
More Information
Acknowledgment
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on the National Centers for Biomedical Computing can be obtained from http://nihroadmap.nih.gov/bioinformatics
References
Publications related to this module go here. Links to pdfs would be useful.