Difference between revisions of "Slicer3.4:Training"
From Slicer Wiki
| Line 40: | Line 40: | ||
|} | |} | ||
| + | = Slicer Tutorial Contest (under construction)= | ||
| + | |||
| + | The following tutorials were part of the [http://www.na-mic.org/Wiki/index.php/Events:TutorialContestJune2009Summer 2009 Slicer tutorial contest]: | ||
| + | |||
| + | * [http://www.na-mic.org/Wiki/images/4/4d/Microscopy-Confocal-TrainingTutorial-2009JUNE.pdf Confocal Microscopy] '''(First Prize)''' | ||
| + | * [http://www.na-mic.org/Wiki/index.php/File:ARCTIC-Slicer3-Tutorial.pdf ARCTIC: Automatic Regional Cortical ThICkness] '''(Second Prize)''' | ||
| + | * [http://www.na-mic.org/Wiki/images/0/06/DBP2JohnsHopkinsTransRectalProstateBiopsy_TutorialPres2009June.pdf Trans-rectal MR guided prostate biopsy] | ||
| + | * [http://www.na-mic.org/Wiki/index.php/File:Stochastic_June09_1.ppt Python Stochastic Tractography Module] | ||
| + | * [http://www.na-mic.org/Wiki/images/0/06/DBP2JohnsHopkinsTransRectalProstateBiopsy_TutorialPres2009June.pdf White Matter Lesion Classification] | ||
= Older Versions = | = Older Versions = | ||
*Visit the [http://wiki.na-mic.org/Wiki/index.php/Slicer3.2:Training Slicer 3.2 training page]. | *Visit the [http://wiki.na-mic.org/Wiki/index.php/Slicer3.2:Training Slicer 3.2 training page]. | ||
*Visit the [http://wiki.na-mic.org/Wiki/index.php/Slicer:Workshops:User_Training_101 Slicer 2 training page]. | *Visit the [http://wiki.na-mic.org/Wiki/index.php/Slicer:Workshops:User_Training_101 Slicer 2 training page]. | ||
Revision as of 20:20, 19 August 2009
Home < Slicer3.4:TrainingContents
Software Installation
- The Slicer download page contains information on how to obtain a compiled version of Slicer for a variety of platforms and where to find the source code for Slicer 3.
Software Documentation
- For the Slicer 3.4 manual pages please click here. These pages are the reference manual for Slicer 3.4 and briefly explain the functionality found in panels and modules.
Slicer 3.4 Tutorials
- The following table contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment to accomplish certain tasks.
| Category | Tutorial | Sample Data | |
| Basic | Slicer3Minute Tutorial The Slicer3Minute tutorial is an introduction to the advanced 3D visualization capabilities of Slicer3.4. Audience: First time users. |
Slicer3Minute dataset The Slicer3Minute dataset contains an MR scan of the brain and 3D reconstructions of the anatomy |

|
| Core | Slicer3Visualization Tutorial The Slicer3Visualization tutorial guides through 3D data loading and visualization in Slicer3.4. Audience: All beginning users including clinicians, scientists, engineers and programmers. |
Slicer3Visualization dataset The Slicer3VisualizationDataset contains two MR scans of the brain, a pre-computed labelmap and 3D reconstructions of the anatomy. |
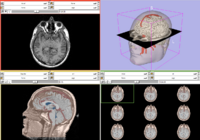
|
| Core | Programming in Slicer3 Tutorial The Programming in Slicer3 tutorial is an introduction to the the integration of stand-alone programs outside of the Slicer3 source tree. Audience: Programmers and Engineers. |
HelloWorld_Plugin.zip The HelloWorld tutorial dataset contains an MR scan of the brain and pre-computed xml and C++ files for integrating the Hello World plug-in to Slicer3. |
|
| Specialized | 3D Visualization of FreeSurfer Data The course guides through 3D visualization of FreeSurfer brain segmentations, surface reconstruction and parcellation results in Slicer3.4. Audience: All users. |
FreeSurferTutorialData.zip The FreeSurfer dataset contains an MR scan of the brain and pre-computed FreeSurfer segmentation and cortical surface reconstructions. |
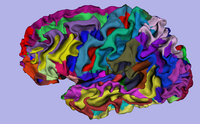
|
Slicer Tutorial Contest (under construction)
The following tutorials were part of the 2009 Slicer tutorial contest:
- Confocal Microscopy (First Prize)
- ARCTIC: Automatic Regional Cortical ThICkness (Second Prize)
- Trans-rectal MR guided prostate biopsy
- Python Stochastic Tractography Module
- White Matter Lesion Classification
Older Versions
- Visit the Slicer 3.2 training page.
- Visit the Slicer 2 training page.