Difference between revisions of "Slicer3:VisualBlog"
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<gallery caption="2009" widths="200px" perrow="4"> | <gallery caption="2009" widths="200px" perrow="4"> | ||
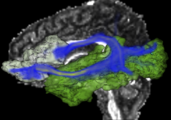
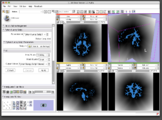
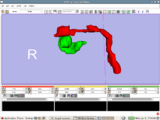
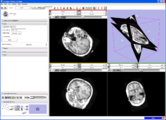
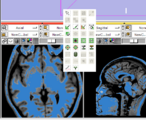
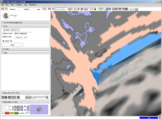
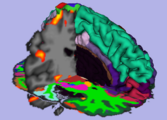
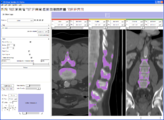
| + | Image:Stochastic-Tractography-01040-lh-all-3D-cropped.png|[http://www.na-mic.org/Wiki/index.php/Summer2009:VCFS ''New Stochastic Tractography''] tools for analysis of diffusion volumes implemented as Python modules by Julien von Seibenthal. | ||
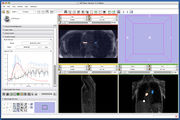
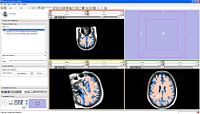
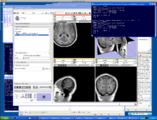
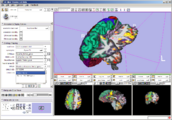
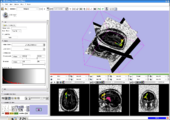
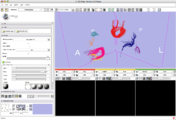
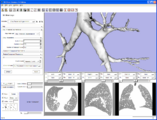
Image:3DSlicerFourDAnalysis Screenshot.png |[http://www.na-mic.org/Wiki/index.php/2009_Summer_Project_Week_4D_Imaging '''Four Dimensional Image (Time Series) Analysis'''] Junichi Tokuda has implemented a slicer module for dynamic MR analysis and a general framework for working with time series volume groups. | Image:3DSlicerFourDAnalysis Screenshot.png |[http://www.na-mic.org/Wiki/index.php/2009_Summer_Project_Week_4D_Imaging '''Four Dimensional Image (Time Series) Analysis'''] Junichi Tokuda has implemented a slicer module for dynamic MR analysis and a general framework for working with time series volume groups. | ||
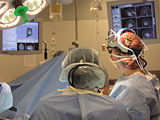
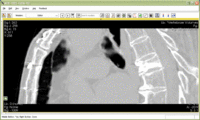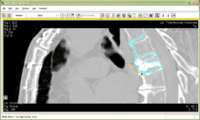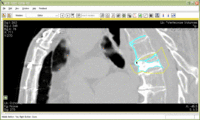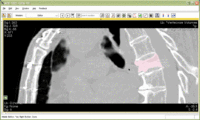
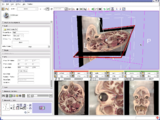
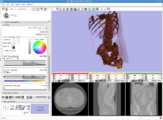
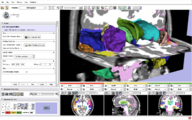
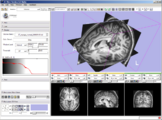
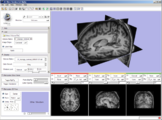
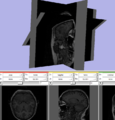
Image:Slicer dti seeding or.jpg|[http://www.na-mic.org/Wiki/index.php/2009_Summer_Project_Week_Slicer3_Brainlab_Introduction '''Intraoperative navigation experiments'''] integrate slicer3 with the commercial [http://www.brainlab.com/scripts/website_english.asp BrainLab] system and Yale's [http://bioimagesuite.org/ BioImageSuite] software. | Image:Slicer dti seeding or.jpg|[http://www.na-mic.org/Wiki/index.php/2009_Summer_Project_Week_Slicer3_Brainlab_Introduction '''Intraoperative navigation experiments'''] integrate slicer3 with the commercial [http://www.brainlab.com/scripts/website_english.asp BrainLab] system and Yale's [http://bioimagesuite.org/ BioImageSuite] software. | ||
Revision as of 21:03, 20 July 2009
Home < Slicer3:VisualBlog- 2009
New Stochastic Tractography tools for analysis of diffusion volumes implemented as Python modules by Julien von Seibenthal.
Four Dimensional Image (Time Series) Analysis Junichi Tokuda has implemented a slicer module for dynamic MR analysis and a general framework for working with time series volume groups.
Intraoperative navigation experiments integrate slicer3 with the commercial BrainLab system and Yale's BioImageSuite software.
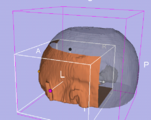
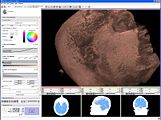
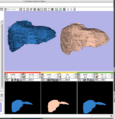
New skull stripping extension module implemented by Xiaodong Tao.
CompareView for Longitudinal Visualization of MS implemented by Jim Miller and demonstrated using a set of 5 scans of a patient with multiple sclerosis. Data provided by Dominik Meier of the Center for Neurological Imaging
- 2008
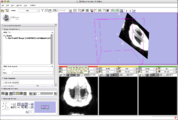
Constrained Level Tracing Algorithm implemented by Andrew Li and his team at Synarc built on the Slicer3 Editor module has been deployed for bone density measurements in support of clinical trials. Synarc employees participated in the 2008 Slicer Training event at Stanford University.
Start frame from time lapse movie showing "Chameleon Tumor" progression. Video courtesy by Ervin Berenyi and Andras Jakab, Department of Medical Laboratory and Diagnostic Imaging, University of Debrecen Medical School and Health Science Center.
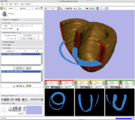
Image showing the IA-FEMesh module in Slicer3.
This image shows the display of material properties assigned to a hexahedral mesh. This image was created by Curl Lisle, Nicole Grosland, Kiran Shivanna, Steve Pieper, and Vincent MagnottaDistance Transform from a label map calculated by Steve Pieper using the NumpyScript module created by Luca Antiga. See Slicer3:Python for more info.
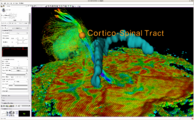
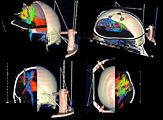
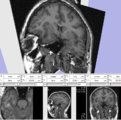
Exploring DTI visualization using the Neurosurgical Tutorial. A virtual probe (fiducial) was positioned in the motor cortex and used as a seed for tractography. The region adjacent to the tumor was used as a seeding area for a separate tractography, which is represented as thin lines with ellipsoid glyphs.
Color Image Reslice Demonstrates the ability to read a sequence of jpg files as a color volume and apply 3D interactive reslice tools. Image data is Visible Human 2 courtesy of the NAC Clinical Computational Anatomy Core
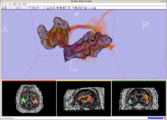
Planning for CT Radiosurgery augmented with diffusion tractography (fibers: internal capsule, corpus callosum and the environment of the tumor, which is a metastatic tumor in the parietal lobe) Image Courtesy of University of Debrecen, Medical School and Health Science Center more information...
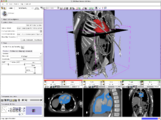
Cardiac segmentation and CT Volume Rendering, February 2008
Using data and segmentations from the collaboration with Boston Children's Hospital Pediatric Cardiology.Partial image update using OpenIGTLink, February 2008
OpenIGTLink allows an external imaging scanner (Ultrasound/CT/MR) to update part of a volume, which has already been loaded on the Slicer. This is helpful if the imaging plane sweeps the subject, and images are transfered to the Slicer on-the-fly.The First MRML node rendering using CUDA February 2008
Cuda Volume Rendering provides a Volume Rendering Method on the new and advanced NVidia Hardware.The First Implementation of OpenIGTLink, January 2008
OpenIGTLink protocol provides plug-and-play connectivity to tracking devices, imagers (MR, ultrasound ...) and other medical devices. A real-time image transfer to the Slicer in 10 fps is demonstrated in the screenshot.Image from Haiying Liu, Patrick Cheng, Noby Hata, Junichi Tokuda, Luis Ibanez, and Steve Pieper, January 2008.
In the NAMIC All Hands Meeting 2008, the bi-directional socket communication has been established between the Tracker Daemon in Slicer 3 and IGSTK server which acquires tracking data from device and sends it to Tracker Daemon. The speed of communication is controlled by Slicer.
- 2007
Image from Steve Pieper, Luis Ibanez, Haiying Liu December 2007
Result of Slicer for IGT workshop showing Slicer3 transform node being updated by a IGSTK tracker process.Image from Steve Pieper, December 2007
Display of segmented heart data results using volume rendering inside Slicer3. See JHU Computational Anatomy Portal for more information.Image from Tri Ngo and Steve Pieper, November 2007
Display of Stochastic Tractography results using volume rendering inside Slicer3. See here for more information.Image from Andy Freudling on October 2007
First results of volume rendering inside Slicer3. See here for more information.Image from Dirk, Matthew, Jim, and Steve on Monday, August 13, 2007
A new label map smoothing tool has been added to help with our collaboration with Children's Hospital Boston, SCI at University of Utah and Northeastern University. The unfiltered labelmap is shown in blue, and the filtered results are shown in peach.Image from Brad and Kilian on Wednesday, June, 21, 2007
Example of EMSegmenter in Slicer3Image from pieper on Friday, June, 9, 2007
Slicer3 Module for Stochastic Tractography from MIT (Ngo, Golland) and BWH (Westin, Kubicki).
- 2006
Image from wjp on Friday, December 29, 2006 at 3:23PM
Module choose and navigation functionality integrated into the application toolbar to create more space for module GUIs on the side panel. Slice Controller widgets also updated to have more functionality available, a button to link and unlink their control, and an improved visual design.Image from pieper on Tuesday, August 08, 2006 at 4:26PM
A number of things have come together to allow the new prototype editor module. More ...Image from pieper on Thursday, July 27, 2006 at 5:21PM
Shows that a single pixel voxel in black is exactly bounded by a unit cube in RAS space. Also the blinking eye buttons for slice visiblility are hooked up and there is now a red-yellow-green color coding for the slice windows (carried over from slicer2).Image from pieper on Thursday, July 13, 2006 at 4:49PM
Test of interactive slice textured plane control. Not yet eneabled through GUI, but driven by a test script that you can source into the console. More ...Image from pieper on Monday, July 03, 2006 at 3:35PM
Test of the new logo widget for adding module-specific watermark logos directly in the 3d view. Uses a vtkLogoWidget from VTK cvs head. Kitware is working on cmake support to notify developers when a VTK cvs update is needed. Once that is done, the logo code will be added to slicer3 svn.
About the VisualBlog
The VisualBlog is meant to be an easy place to upload screenshot so that both developers and outside observes can track the progress of the project.
Many thanks to developers and users for contributing their images here.