Difference between revisions of "Slicer3:Module:Fuzzy Surface Creation-Documentation"
| Line 91: | Line 91: | ||
For the following example an image dataset of a human head was used. | For the following example an image dataset of a human head was used. | ||
| − | <gallery perrow="1" heights="200" widths=" | + | <gallery perrow="1" heights="200" widths="650"> |
Image:PrivacyOriginalData2D.png|Original dataset before manipulation | Image:PrivacyOriginalData2D.png|Original dataset before manipulation | ||
Image:PrivacyFuzzyData2D.png|dataset after manipulation | Image:PrivacyFuzzyData2D.png|dataset after manipulation | ||
| Line 98: | Line 98: | ||
After execution of the privacy module there is a additional surface visible on the front of the head surface. | After execution of the privacy module there is a additional surface visible on the front of the head surface. | ||
| − | <gallery heights=" | + | <gallery heights="250" widths="200"> |
Image:PrivacyOriginalDataSlice.png|Original | Image:PrivacyOriginalDataSlice.png|Original | ||
Image:PrivacyFuzzyDataSlice.png|Manipulated | Image:PrivacyFuzzyDataSlice.png|Manipulated | ||
Revision as of 18:27, 4 September 2008
Home < Slicer3:Module:Fuzzy Surface Creation-DocumentationReturn to Slicer Documentation
Fuzzy Surface Creation
General Information
Data sharing became a big issue in the last few years. Unfortunately for research data sharing is a difficult undertaking because of patient privacy protection. This constricts the exchange of patient secific data between different research groups and faculties.
MRI and CT data itself wouldn't be that delicate since the identification of a patient isn't obvious. But the rendering of the data into a 3D view reveals a lot of information about the skin and skull structure of a patient and therefore makes a patient easily identifiable.
The aim of this module is therefore to hide as much surface information of the patient as possible but also to keep internal structures untouched.
Module Type & Category
Type: CLI
Category: Privacy Protection
Authors, Collaborators & Contact
- Author: Bernhard Pflug, University for medical informatics and technology, Tyrol, Austria
- Contact: Bernhard Pflug, bernhard.pflug@gmail.com
Module Description
The purpose of this module is to hide as much surface details of a patient as possible. Therefore it adds an additional surface to the front of the object in the image depending on different calulated parameters.
As a first step the module detects the object in the image. Subsequent it adds additional data on the detected surface depending on several calculated parameters. As a result it creates an additional surface on the object which hides skin and skull details so that patient identification isn't obvious any more.
Usage
This module is a CLI Module for Slicer. It is mainly seperated into two parts:
- Preprocessing
- Fuzzy Surface Creation
Preprocessing
The first step is meant to be the pre-processing step. The Otsu - Threshold - Filter is used as it provides quite good segmentation results on the object in the image and is considerably robust against noise, especially if it is not connected to the object.
The pre-processing task defines the following parameters:
- bright objects
- number of histogram bins
- face connected
- minimum object size
For further information about these parameters check the Otsu - Threhold - Filter in Slicer3.
Fuzzy Surface Creation
The second step is the actual creation of the fuzzy surface. Important to mention is that this algorithm works local, so it creates the additional surface for each voxel without considering the environment!
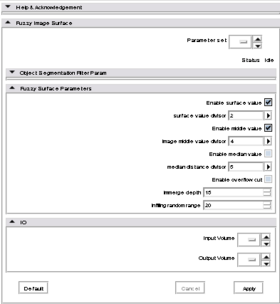
It defines the following parameters:
- Surface value
- Enable surface value
- Surface value divisor
- Middle value
- Enable middle value
- Middle value divisor
- Median value
- Enable median value
- Median value divisor
- Enable overflow cut
- Infiltration depth
- Infilling random range
- Surface-, Middle-, Median value
- These parameters are calculated and used to compute the height of the added surface. They define the consideration of the calculated value and there aggregation to the final additional height.The parameters serve as divisors, so the lower they are set the more they are considered. The surface parameter defines the distance of a surface point to the border of the image, the middle value the distance from the point to the center of the z-axis and the median value the distance between the point and the median of all surface values of the current slice. Furthermore every value can be en- or disabled by the respective check box.
- Enable overflow cut
- By checking the overflow cut box every value above the created additional surface is set to zero. This improves the fuzzy effect because especially values which are not detected by the Otsu Threhold Preprocessing like especially skin which wouldn't be influenced otherwise can be removed. But this also means that all information above the created surface will be lost.
- Infiltration Depth
- The infiltration depth defines the number of voxels the algorithm goes into the detected surface structure to overlap original voxels with the new created ones. The deeper the algorithm goes into the object the more information will be lost.
- Infilling random range
- For the intensity of the added voxels an average value of the underneath voxels (5 by default) from the surface value is determined. For adding the new surface this value including a random created value is used for intensity estimation. The random range parameter defines the range for the random created value.
Example
For the following example an image dataset of a human head was used.
After execution of the privacy module there is a additional surface visible on the front of the head surface.
This example shows the data in the 3D view in slicer before and after the usage of two different parameter settings in the module.
From another point of view:
Development
This module is splitted into two C++ files:
- itkFuzzySurface
- This class is a subclass of itkImageToImageFilter and handles the fuzzy surface creation. It's a fully functional itk filter and can be embedded in every appropriate itk image processing pipeline. This structure makes the algorithm abstract from it's preprocessing, provides better separation between functional independent parts and offers better maintenance and reusability.
- Privacy
- This file includes the main function and therefore the execution of the module. It first calls the preprocessing steps (Otsu Threshold) and afterwards the fuzzySurfaceFilter. Additionally all required parameters for the algorithms are set.
Source code & Documentation
Except this wiki page there is documentation available in the source code. I tried to structure the files as good as possible and also to document any necessary part. I did this especially for people who are not really familiar with ITK, generic programming and Slicer. Therefore the files may seem a little bit overdocumented for sophisticated developers.
More Information
Acknowledgment
The development took place in the Surgical Planning Laboratory, Harvard Medical School and Brigham and Women's Hospital, under the supervision of Mr. Steve Pieper Ph.D.