Difference between revisions of "Documentation/4.10/Modules/DoseComparison"
(Nightly -> 4.10) |
m (Text replacement - "slicerWiki/index.php" to "wiki") |
||
| Line 12: | Line 12: | ||
Contributor: Greg Sharp (Massachusetts General Hospital)<br> | Contributor: Greg Sharp (Massachusetts General Hospital)<br> | ||
Contact: Csaba Pinter, <email>pinter@cs.queensu.ca</email><br> | Contact: Csaba Pinter, <email>pinter@cs.queensu.ca</email><br> | ||
| − | [http://www.slicer.org/ | + | [http://www.slicer.org/wiki/Documentation/Nightly/Extensions/SlicerRT Back to SlicerRT home] |
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
{{documentation/{{documentation/version}}/module-introduction-logo-gallery | {{documentation/{{documentation/version}}/module-introduction-logo-gallery | ||
| Line 36: | Line 36: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Tutorials}} | {{documentation/{{documentation/version}}/module-section|Tutorials}} | ||
| − | See [http://www.slicer.org/ | + | See [http://www.slicer.org/wiki/Documentation/Nightly/Extensions/SlicerRT#Tutorials SlicerRT main page] |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 57: | Line 57: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Similar Modules}} | {{documentation/{{documentation/version}}/module-section|Similar Modules}} | ||
| − | [http://www.slicer.org/ | + | [http://www.slicer.org/wiki/Documentation/Nightly/Modules/SegmentComparison Segment Comparison] |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 17:05, 21 November 2019
Home < Documentation < 4.10 < Modules < DoseComparison
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work is part of the SparKit project, funded by An Applied Cancer Research Unit of Cancer Care Ontario with funds provided by the Ministry of Health and Long-Term Care and the Ontario Consortium for Adaptive Interventions in Radiation Oncology (OCAIRO) to provide free, open-source toolset for radiotherapy and related image-guided interventions. | |||||||
|
Module Description
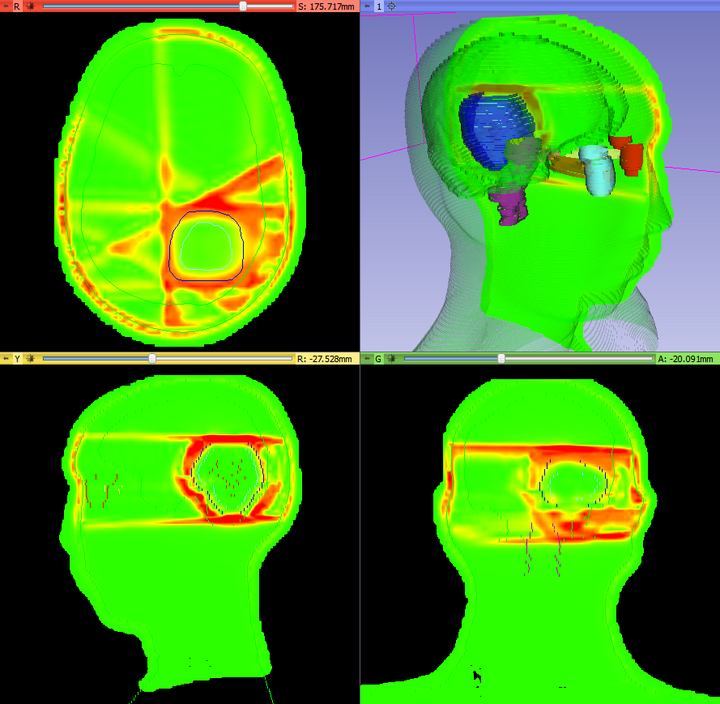
The DoseComparison module computes the difference between two co-registered dose volumes using the gamma dose distribution comparison method. This module uses the gamma Plastimatch algorithm internally.
Use Cases
Comparing dose distributions
Tutorials
Panels and their use
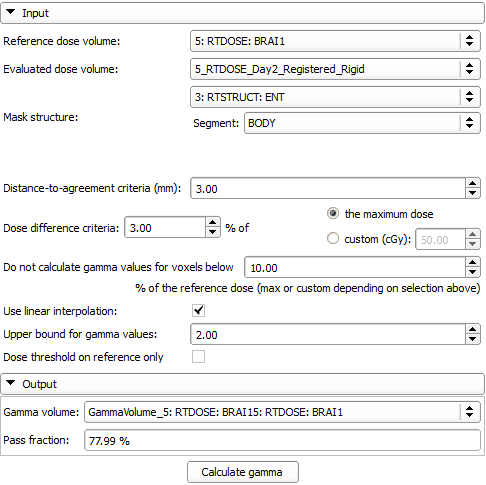
- Reference dose volume: Dose distribution used as reference
- Evaluated dose volume: Dose distribution used as evaluation/compare
- Mask structure: Segmentation and contained segment within which the gamma index is calculated, but not outside (it will be 0)
- Distance-to-agreement (DTA): Mark dose voxel as failed if closest dose voxel with same value found in distance higher than this threshold, see Low paper below
- Dose difference criteria: Mark dose voxel as failed if dose difference is equal or greater than this threshold, see Low paper below
- Maximum value / Custom: Reference dose value for threshold can be either the maximum dose in the dose map, or a custom cGy value
- Do not calculate gamma values for voxels below analysis threshold
- Use linear interpolation
- Upper bound for gamma values: If gamma value would be higher than this value, the value will be set instead
- Dose threshold on reference only
- Gamma volume: Output volume to contain gamma map. Needs to be created using the combobox. Name will be automatically filled from the names of the input volumes
Similar Modules
References
- Low, D. A.; Harms, W. B.; Mutic, S. & Purdy, J. A. A technique for the quantitative evaluation of dose distributions Med. Phys, 1998, 25 (5), 656/6
- Low, D. A. & Dempsey, J. F. Evaluation of the gamma dose distribution comparison method Med. Phys, 2003, 30 (9), 2455/10
Information for Developers
N/A