|
|
| Line 250: |
Line 250: |
| | |} | | |} |
| | | | |
| − | = 3D Slicer Tutorial contests= | + | = 3D Slicer version 4.7 Tutorial Contest= |
| − | | |
| − | ==Winter 2017 Tutorial contest==
| |
| | | | |
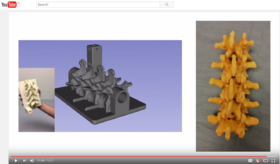
| | ===Segmentation for 3D printing=== | | ===Segmentation for 3D printing=== |
| Line 313: |
Line 311: |
| | |align="right"| | | |align="right"| |
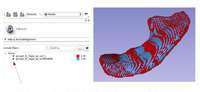
| | [[File:SlicerWinterProjectWeek2017-FiberBundleVolumeMeasurements.png | 200px]]. | | [[File:SlicerWinterProjectWeek2017-FiberBundleVolumeMeasurements.png | 200px]]. |
| − | |}
| |
| − |
| |
| − | ==Winter 2016 Tutorial contest==
| |
| − |
| |
| − | ===Subject Hierarchy===
| |
| − | {|width="100%"
| |
| − | |
| |
| − | *The [http://wiki.na-mic.org/Wiki/images/2/27/SubjectHierarchy.TutorialContestWinter2016.pdf Subject Hierarchy] tutorial demonstrates the basic usage and potential of Slicer’s data manager module Subject Hierarchy using a two-timepoint radiotherapy phantom dataset.
| |
| − | *Author: Csaba Pinter, Queen's University, Canada
| |
| − | *Dataset: [http://slicer.kitware.com/midas3/download/item/205404/SlicerRT_WorldCongress_TutorialIGRT_Dataset.zip SlicerRT_WorldCongress_TutorialIGRT_Dataset] The tutorial dataset is a two-timepoint phantom dataset taken from a RANDO head&neck phantom. It contains two studies, the planning one is a DICOM study consisting of a CT grayscale image and radiotherapy data: contours, dose distribution, treatment beams, plan information. The second timepoint consists of a CT NRRD volume and a dose NRRD volume.
| |
| − | |align="right"|
| |
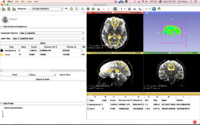
| − | [[File:SubjectHierarchyTutorial.png | 200px]].
| |
| − | |}
| |
| − |
| |
| − | ===Fiber Bundle Selection and Scalar Measurements===
| |
| − | {|width="100%"
| |
| − | |
| |
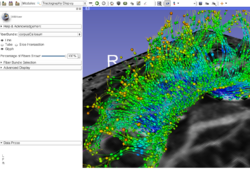
| − | *The [https://github.com/SlicerDMRI/slicerdmri.github.io/raw/master/docs/tutorials/FiberBundleSelectionAndScalarMeasurement.pdf Fiber Bundle Selection and Scalar Measurements] tutorial guides through the use of the Diffusion Bundle Selection module and the Fiber Tract Scalar Measurement module for diffusion MRI tractography data analysis.
| |
| − | *Author: Fan Zhang, University of Sydney Australia and Brigham and Women's Hospital
| |
| − | *Dataset: [[media:FiberBundleSelectionAndScalarMeasurement_TutorialContestWinter2016.zip| Fiber Bundle Selection And Scalar Measurement Tutorial Dataset]]
| |
| − | |align="right"|
| |
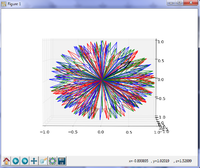
| − | [[File:FiberBundleSelectionAndScalarMeasurement_TutorialContestWinter2016_Snapshot.png|200px]]
| |
| − | |}
| |
| − |
| |
| − | ===Plastimatch ===
| |
| − | {|width="100%"
| |
| − | |
| |
| − | *The [http://www.na-mic.org/Wiki/images/5/5c/Plastimatch_TutorialContestWinter2016.pdf Plastimatch tutorial] guides through registration and wrapping of DICOM and DICOM-RT data using the Plastimatch extension of 3D Slicer.
| |
| − | *Author: Gregory Sharp, Massachusetts General Hospital
| |
| − | *Dataset: [http://www.na-mic.org/Wiki/index.php/File:Plastimatch_TutorialContestWinter2016.zip Plastimatch Tutorial Dataset]
| |
| − | |align="right"|
| |
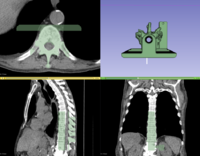
| − | [[File:PlastimatchTutorial_Winter2016Contest.png|200px]]
| |
| − | |}
| |
| − |
| |
| − | ===UKF ===
| |
| − | {|width="100%"
| |
| − | |
| |
| − | *The [https://github.com/SlicerDMRI/slicerdmri.github.io/raw/master/docs/tutorials/UKFTractography.pdf UKF tutorial] guides through the use of the Unscented Kalman Filter (UKF) tractography module.
| |
| − | *Author: Pegah Kahali, Brigham and Women's Hopital
| |
| − | *Dataset: [http://www.na-mic.org/Wiki/index.php/File:UKF-Tractography_TutorialContestWinter2016.zip UKF tutorial Dataset]
| |
| − | |align="right"|
| |
| − | [[File:UKF_Winter2016.png|200px]]
| |
| − | |}
| |
| − |
| |
| − | ==Summer 2014 Tutorial contest==
| |
| − |
| |
| − | ===Cardiac Agatston Tutorial===
| |
| − | {|width="100%"
| |
| − | |
| |
| − | *[http://wiki.na-mic.org/Wiki/index.php/File:TutorialContest_CardiacAgatstonScoring_2014.pdf Cardiac Agatston Scoring Tutorial]
| |
| − | *Authors: Jessica Forbes, Hans Johnson, University of Iowa
| |
| − | *Dataset: [http://wiki.na-mic.org/Wiki/index.php/File:CardiacAgatstonMeasures_TutorialContestSummer2014.zip Cardiac Agatston Scoring Tutorial Dataset]
| |
| − | |align="right"|
| |
| − | [[File:CardiacAgatstonMeasuresModuleScreenshot.jpg| 250px]]
| |
| − | |}
| |
| − |
| |
| − | ===CMR Toolkit LA workflow===
| |
| − | {|width="100%"
| |
| − | |
| |
| − | *[http://wiki.na-mic.org/Wiki/index.php/File:CMRToolkitLAWorkflow_TutorialContestSummer2014.pdf CMR Toolkit LA Workflow Tutorial]
| |
| − | *Authors: Salma Bengali, Josh Cates, University of Utah
| |
| − | *Dataset: [http://wiki.na-mic.org/Wiki/index.php/File:CMRToolkitLAWorkflowData_TutorialContestSummer2014.zip CMRToolkitLAWorkflow Dataset]
| |
| − | |align="right"|
| |
| − | [[Image:Utah_SummerContest2014_tutorial.png|300px]]
| |
| − | |}
| |
| − |
| |
| − | ==Summer 2013 Tutorial contest==
| |
| − |
| |
| − | ===Cardiac MRI Toolkit===
| |
| − | {|width="100%"
| |
| − | |
| |
| − | *[[Media:Cardiac MRI Toolkit Tutorial Summer2013.pdf|Cardiac MRI Toolkit]]
| |
| − | *Authors: Salma Bengali, Josh Cates, SCI, Utah
| |
| − | *Dataset: [[Media:Cardiac_MRI_Toolkit_Tutorial_Data.zip|Cardiac MRI Toolkit Tutorial Dataset]]
| |
| − | |align="right"|
| |
| − | [[Image:CMRToolkit_Tutorial_Image.png|250px]]
| |
| − | |}
| |
| − |
| |
| − | ===HelloCLI===
| |
| − | {|width="100%"
| |
| − | |
| |
| − | *[[Media:Hello_CLI_TutorialContestSummer2013.pdf|HelloCLI]]
| |
| − | *Authors: Nadya Shusharina, Greg Sharp, MGH, Boston
| |
| − | *Dataset: [[Media:Hello_CLI_TutorialContestSummer2013.zip|HelloCLI Dataset]]
| |
| − | |align="right"|
| |
| − | [[Image:Cli_icon.png|300px]]
| |
| − | |}
| |
| − |
| |
| − | ===SlicerRT===
| |
| − | {|width="100%"
| |
| − | |
| |
| − | *[[Media:SlicerRT_TutorialContestSummer2013.pdf|SlicerRT Tutorial]]
| |
| − | *Authors: Csaba Pinter, Andras Lasso (Queen's), Kevin Wang (PMH, Toronto)
| |
| − | *Dataset: [[Media:CsabaPinter-SlicerRtTutorial_Namic2013June.zip|SlicerRT Dataset]]
| |
| − | |align="right"|
| |
| − | [[Image:667px-SlicerRT_0.10_IsocenterShiftingEvaluation.png|250px]]
| |
| − | |}
| |
| − |
| |
| − | ===DTIPrep===
| |
| − | {|width="100%"
| |
| − | |
| |
| − | *[[Media:DTIPrep_TutorialContestSummer2013.pdf|DTIPrep]]
| |
| − | *Authors: Dave Welch, SINAPSE, IOWA
| |
| − | *Dataset: [[Media:DTIPrepData_TutorialContestSummer2013.zip|DTIPrep Dataset]]
| |
| − | |align="right"|
| |
| − | [[Image:DTIPrep-tutorial.png|250px]]
| |
| − | |}
| |
| − |
| |
| − | == Summer 2012 Tutorial contest ==
| |
| − |
| |
| − | ===Automatic Left Atrial Scar Segmenter ===
| |
| − | {|width="100%"
| |
| − | |
| |
| − | *[http://wiki.na-mic.org/Wiki/index.php/CARMA-LA-Scar_TutorialContestSummer2012 Automatic Left Atrial Scar Segmenter]
| |
| − | *Authors: Greg Gardner, Josh Cates, SCI, Utah
| |
| − | *Dataset: [http://wiki.na-mic.org/Wiki/index.php/File:CARMA-LA-Scar_TutorialContestSummer2012.zip CARMA-LA-Scar data]
| |
| − | |align="right"|
| |
| − | [[Image:Carma afib auto scar.png|250px]]
| |
| − | |}
| |
| − |
| |
| − | ===Qualitative and quantitative comparison of two RT dose distributions===
| |
| − | {|width="100%"
| |
| − | |
| |
| − | *[http://www.na-mic.org/Wiki/index.php/File:PlastimatchDose_TutorialContestSummer2012.pdf Qualitative and quantitative comparison of two RT dose distributions]
| |
| − | *Authors: James Shackleford, Nadya Shusharina, Greg Sharp, MGH
| |
| − | |align="right"|
| |
| − | [[Image:PlastimatchDose.png|250px]]
| |
| − | |}
| |
| − |
| |
| − | ===Dose accumulation for adaptive radiation therapy===
| |
| − | {|width="100%"
| |
| − | |
| |
| − | *[http://www.na-mic.org/Wiki/index.php/File:DoseAccumulationforAdaptiveRadiationTherapy_TutorialContestSummer2012.pdf Dose accumulation for adaptive radiation therapy]
| |
| − | *Authors: Kevin Wang, Csaba Pinter, Andras Lasso, PMH, Queen's
| |
| − | |align="right"|
| |
| − | [[Image:AdaptiveradiationTherapy.png|250px]]
| |
| − | |}
| |
| − |
| |
| − | ===WebGL Export===
| |
| − | {|width="100%"
| |
| − | |
| |
| − | *[http://www.na-mic.org/Wiki/index.php/File:WebGLExport_TutorialContestSummer2012.pdf WebdGLExport]
| |
| − | *Authors: Nicolas Rannou, Daniel Haehn, Children's Hospital
| |
| − | |align="right"|
| |
| − | [[Image:WebGLExport.png|250px]]
| |
| − | |}
| |
| − |
| |
| − | ===OpenIGTLink===
| |
| − | {|width="100%"
| |
| − | |
| |
| − | *[http://wiki.slicer.org/slicerWiki/images/f/f1/OpenIGTLinkTutorial_Slicer4.1.0_JunichiTokuda_Apr2012.pdf OpenIGTLink]
| |
| − | *Authors: Junichi Tokuda, BWH
| |
| − | |align="right"|
| |
| − | [[Image:OpenIGTLink.png|250px]]
| |
| | |} | | |} |
| | | | |