Difference between revisions of "Documentation/Nightly/Modules/LSSegmenter"
From Slicer Wiki
Acsenrafilho (talk | contribs) (Created module's wiki page) |
Acsenrafilho (talk | contribs) m |
||
| Line 22: | Line 22: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Module Description}} | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
| − | This module offer a | + | This module offer a voxel-intensity lesion segmentation method based on logistic contrast enhancement and threshold level. At moment, this method was studied on hyperintense T2-FLAIR lesion segmentation in Multiple Sclerosis lesion segmentation. |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 28: | Line 28: | ||
* Use Case 1: | * Use Case 1: | ||
**Something. | **Something. | ||
| − | <gallery widths=" | + | <gallery widths="300px" heights="300px" perrow="3"> |
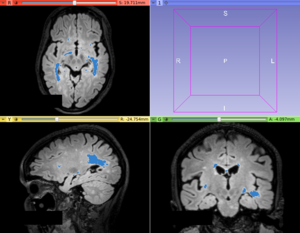
| − | Image: | + | Image:T2FLAIR_patient.png|Input T2-FLAIR image with hyperintense Multiple Sclerosis lesions |
| − | Image: | + | Image:T2FLAIR_patient_lesionLabel.png|Lesion map resulted from LS Segmenter module |
</gallery> | </gallery> | ||
| Line 39: | Line 39: | ||
[[Image:lssegmenter_gui.png|thumb|380px|User Interface]] | [[Image:lssegmenter_gui.png|thumb|380px|User Interface]] | ||
IO: | IO: | ||
| − | * | + | *T1 Volume |
| − | ** | + | *T2-FLAIR Volume |
| − | * | + | *Lesion Label |
| − | ** | + | **Output a global lesion mask |
| + | *Is brain extracted? | ||
| + | **Is the input data (T1 and T2-FLAIR) already brain extracted? | ||
| + | *Apply noise attenuation step | ||
| + | **Apply noise attenuation based on the [[Documentation/Nightly/Modules/AADImageFilter|Anisotropic Anomalous Diffusion]] algorithm on the input data (T1 and T2-FLAIR) | ||
| + | *Apply bias field correction step | ||
| + | **Apply bias field correction based on the [http://www.slicer.org/wiki/Documentation/Nightly/Modules/N4ITKBiasFieldCorrection N4ITK Bias Field Correction] algorithm on the input data (T1 and T2-FLAIR) | ||
| − | + | Noise Attenuation Parameters: | |
*Conductance | *Conductance | ||
**The conductance regulates the diffusion intensity in the neighbourhood area. Choose a higher conductance if the input image has strong noise seem in the whole image space. | **The conductance regulates the diffusion intensity in the neighbourhood area. Choose a higher conductance if the input image has strong noise seem in the whole image space. | ||
| − | * | + | *Number Of Iterations |
| + | **The number of iterations regulates the numerical simulation of the anomalous process over the image. This parameters is also related with the de-noising intensity, however it is more sensible to the noise intensity. Choose the higher number of iterations if the image presents high intensity noise which is not well treated by the conductance parameter | ||
| + | *Q Value | ||
| + | **The anomalous parameter (or q value) is the generalization parameters responsible to give the anomalous process approach on the diffusion equation. See the reference paper[1] to choose the appropriate q value (at moment, only tested in MRI T1 and T2 weighted images) | ||
| + | |||
| + | Registration Parameters: (based on [https://www.slicer.org/wiki/Documentation/Nightly/Modules/BRAINSFit BRAINSFit] module) | ||
| + | *Percentage Of Samples | ||
| + | **Percentage of voxel used in registration | ||
| + | *Initiation Method | ||
| + | **Initialization method used for the MNI152 registration | ||
| + | *Interpolation | ||
| + | **Choose the interpolation method used to register the standard space to input image space. Options: Linear, NearestNeighbor, B-Spline | ||
| + | |||
| + | Segmentation Parameters: | ||
| + | *Absolute Error Threshold | ||
| + | **Define the absolute error threshold for gray matter statistics. This measure evaluated the similarity between the MNI152 template and the T2-FLAIR gray matter fluctuation estimative. A higher error gives a higher variability in the final lesion segmentation | ||
| + | *Gamma | ||
| + | **Define the outlier detection based on units of standard deviation in the T2-FLAIR gray matter voxel intensity distribution | ||
| + | *White Matter Matching | ||
| + | **Set the local neighborhood searching for label refinement step. This metric defines the percentage of white matter tissue that surrounds the hyperintense lesions. Higher values defines a conservative segmentation | ||
| + | *Minimum Lesion Size | ||
| + | **Set the minimum lesion size adopted as a true lesion in the final lesion map. Units given in number of voxels | ||
| + | *Gray Matter Mask Value | ||
| + | **Set the mask value that represents the gray matter. Default is defined based on the Basic Brain Tissues module output | ||
| + | *White Matter Mask Value | ||
| + | **Set the mask value that represents the white matter. Default is defined based on the Basic Brain Tissues module output | ||
| + | |||
| + | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Similar Modules}} | {{documentation/{{documentation/version}}/module-section|Similar Modules}} | ||
| − | + | N/A | |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Revision as of 15:43, 9 December 2016
Home < Documentation < Nightly < Modules < LSSegmenter
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: LesionSpotlight | |||||||
|
Module Description
This module offer a voxel-intensity lesion segmentation method based on logistic contrast enhancement and threshold level. At moment, this method was studied on hyperintense T2-FLAIR lesion segmentation in Multiple Sclerosis lesion segmentation.
Use Cases
- Use Case 1:
- Something.
Panels and their use
IO:
- T1 Volume
- T2-FLAIR Volume
- Lesion Label
- Output a global lesion mask
- Is brain extracted?
- Is the input data (T1 and T2-FLAIR) already brain extracted?
- Apply noise attenuation step
- Apply noise attenuation based on the Anisotropic Anomalous Diffusion algorithm on the input data (T1 and T2-FLAIR)
- Apply bias field correction step
- Apply bias field correction based on the N4ITK Bias Field Correction algorithm on the input data (T1 and T2-FLAIR)
Noise Attenuation Parameters:
- Conductance
- The conductance regulates the diffusion intensity in the neighbourhood area. Choose a higher conductance if the input image has strong noise seem in the whole image space.
- Number Of Iterations
- The number of iterations regulates the numerical simulation of the anomalous process over the image. This parameters is also related with the de-noising intensity, however it is more sensible to the noise intensity. Choose the higher number of iterations if the image presents high intensity noise which is not well treated by the conductance parameter
- Q Value
- The anomalous parameter (or q value) is the generalization parameters responsible to give the anomalous process approach on the diffusion equation. See the reference paper[1] to choose the appropriate q value (at moment, only tested in MRI T1 and T2 weighted images)
Registration Parameters: (based on BRAINSFit module)
- Percentage Of Samples
- Percentage of voxel used in registration
- Initiation Method
- Initialization method used for the MNI152 registration
- Interpolation
- Choose the interpolation method used to register the standard space to input image space. Options: Linear, NearestNeighbor, B-Spline
Segmentation Parameters:
- Absolute Error Threshold
- Define the absolute error threshold for gray matter statistics. This measure evaluated the similarity between the MNI152 template and the T2-FLAIR gray matter fluctuation estimative. A higher error gives a higher variability in the final lesion segmentation
- Gamma
- Define the outlier detection based on units of standard deviation in the T2-FLAIR gray matter voxel intensity distribution
- White Matter Matching
- Set the local neighborhood searching for label refinement step. This metric defines the percentage of white matter tissue that surrounds the hyperintense lesions. Higher values defines a conservative segmentation
- Minimum Lesion Size
- Set the minimum lesion size adopted as a true lesion in the final lesion map. Units given in number of voxels
- Gray Matter Mask Value
- Set the mask value that represents the gray matter. Default is defined based on the Basic Brain Tissues module output
- White Matter Mask Value
- Set the mask value that represents the white matter. Default is defined based on the Basic Brain Tissues module output
Similar Modules
N/A
References
- paper
Information for Developers
| Section under construction. |