Difference between revisions of "Documentation/Nightly/Training"
From Slicer Wiki
| Line 194: | Line 194: | ||
Additional (non-curated) videos-based demonstrations using 3D Slicer are accessible on [http://www.youtube.com/results?search_query=3d+slicer&sm=3 You Tube]. | Additional (non-curated) videos-based demonstrations using 3D Slicer are accessible on [http://www.youtube.com/results?search_query=3d+slicer&sm=3 You Tube]. | ||
| + | |||
| + | =Winter 2016 Tutorial contest= | ||
| + | |||
| + | ==Subject Hierarchy== | ||
| + | {|width="100%" | ||
| + | | | ||
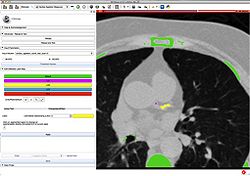
| + | *The [http://wiki.na-mic.org/Wiki/images/2/27/SubjectHierarchy.TutorialContestWinter2016.pdf Subject Hierarchy] tutorial demonstrates the basic usage and potential of Slicer’s data manager module Subject Hierarchy using a two-timepoint radiotherapy phantom dataset. | ||
| + | *Author: Csaba Pinter, Queen's University, Canada | ||
| + | *Dataset: [http://slicer.kitware.com/midas3/download/item/205404/SlicerRT_WorldCongress_TutorialIGRT_Dataset.zip SlicerRT_WorldCongress_TutorialIGRT_Dataset] The tutorial dataset is a two-timepoint phantom dataset taken from a RANDO head&neck phantom. It contains two studies, the planning one is a DICOM study consisting of a CT grayscale image and radiotherapy data: contours, dose distribution, treatment beams, plan information. The second timepoint consists of a CT NRRD volume and a dose NRRD volume. | ||
| + | |align="right"| | ||
| + | [[File:SubjectHierarchyTutorial.png | 200px]]. | ||
| + | |} | ||
| + | |||
| + | ==Fiber Bundle Selection and Scalar Measurements== | ||
| + | {|width="100%" | ||
| + | | | ||
| + | *The [[media:FiberBundleSelectionAndScalarMeasurement_TutorialContestWinter2016.pdf | Fiber Bundle Selection and Scalar Measurements]] tutorial guides through the use of the Diffusion Bundle Selection module and the Fiber Tract Scalar Measurement module for diffusion MRI tractography data analysis. | ||
| + | *Author: Fan Zhang, University of Sydney Australia, Brigham and Women's Hospital | ||
| + | *Dataset: [[media:FiberBundleSelectionAndScalarMeasurement_TutorialContestWinter2016.zip| Fiber Bundle Selection And Scalar Measurement Tutorial Dataset]] | ||
| + | |align="right"| | ||
| + | [[File:FiberBundleSelectionAndScalarMeasurement_TutorialContestWinter2016_Snapshot.png|200px]] | ||
| + | |} | ||
| + | |||
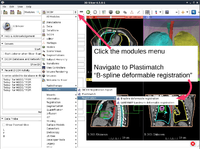
| + | ==Plastimatch == | ||
| + | {|width="100%" | ||
| + | | | ||
| + | *The [http://www.na-mic.org/Wiki/images/5/5c/Plastimatch_TutorialContestWinter2016.pdf Plastimatch tutorial] guides through registration and wrapping of DICOM and DICOM-RT data using the Plastimatch extension of 3D Slicer. | ||
| + | *Author: Gregory Sharp, Massachusetts General Hospital | ||
| + | *Dataset: [http://www.na-mic.org/Wiki/index.php/File:Plastimatch_TutorialContestWinter2016.zip Plastimatch Tutorial Dataset] | ||
| + | |align="right"| | ||
| + | [[File:PlastimatchTutorial_Winter2016Contest.png|200px]] | ||
| + | |} | ||
| + | |||
| + | ==UKF == | ||
| + | {|width="100%" | ||
| + | | | ||
| + | *The [http://www.na-mic.org/Wiki/images/3/3e/UKF-Tractography_TutorialContestWinter2016.pdf UKF tutorial] guides through the use of the Unscented Kalman Filter (UKF) tractography module. | ||
| + | *Author: Pegah Kahali, Brigham and Women's Hopital | ||
| + | *Dataset: [http://www.na-mic.org/Wiki/index.php/File:UKF-Tractography_TutorialContestWinter2016.zip UKF tutorial Dataset] | ||
| + | |align="right"| | ||
| + | [[File:UKF_Winter2016.png|200px]] | ||
| + | |} | ||
=Summer 2014 Tutorial contest= | =Summer 2014 Tutorial contest= | ||
Revision as of 11:57, 12 October 2016
Home < Documentation < Nightly < Training
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction: Slicer Nightly Tutorials
- This page contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment (version Nightly release) to accomplish certain tasks.
- For tutorials for other versions of Slicer, please visit the Slicer training portal.
- For "reference manual" style documentation, please visit the Slicer Nightly documentation page
- For questions related to the Slicer4 Training Compendium, please send an e-mail to Sonia Pujol, Ph.D
- Some of these tutorials are based on older releases of 3D Slicer. The concepts are still useful but bear in mind that some interface elements and features will be different in updated versions.
Contents
- 1 Introduction: Slicer Nightly Tutorials
- 2 General Introduction
- 3 Tutorials for software developers
- 4 Specific functions
- 4.1 Slicer4 Diffusion Tensor Imaging Tutorial
- 4.2 Slicer4 Neurosurgical Planning Tutorial
- 4.3 Slicer4 3D Visualization of DICOM images for Radiology Applications
- 4.4 Slicer4 Quantitative Imaging tutorial
- 4.5 Slicer4 IGT
- 4.6 Slicer4 3D Printing
- 4.7 Slicer4 Image Registration
- 4.8 Subject Hierarchy
- 4.9 Fast GrowCut
- 4.10 Other
- 5 Winter 2016 Tutorial contest
- 6 Summer 2014 Tutorial contest
- 7 Summer 2013 Tutorial contest
- 8 Summer 2012 Tutorial contest
- 9 Additional resources
- 10 External Resources
General Introduction
Slicer Welcome Tutorial
|
Slicer4Minute Tutorial
|
Slicer4 Data Loading and 3D Visualization
|
Tutorials for software developers
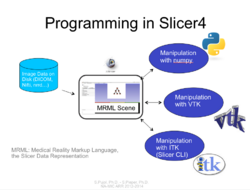
Slicer4 Programming Tutorial
|
For additional Python scripts examples, please visit the Script Repository page
Developing and contributing extensions for 3D Slicer
|
Specific functions
Slicer4 Diffusion Tensor Imaging Tutorial
|
Slicer4 Neurosurgical Planning Tutorial
|
Slicer4 3D Visualization of DICOM images for Radiology Applications
|
Slicer4 Quantitative Imaging tutorial
|
Slicer4 IGT
|
Slicer4 3D Printing
|
|
Slicer4 Image Registration
|
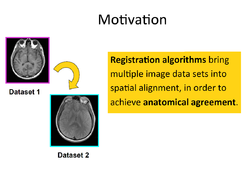
|
- Based on: 3D Slicer version 4.5
See the Registration Library for worked out registration examples with data.
Subject Hierarchy
|
Fast GrowCut
|

|
Other
Additional (non-curated) videos-based demonstrations using 3D Slicer are accessible on You Tube.
Winter 2016 Tutorial contest
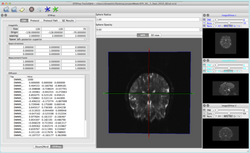
Subject Hierarchy
|
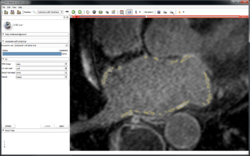
Fiber Bundle Selection and Scalar Measurements
|
Plastimatch
|
UKF
|
Summer 2014 Tutorial contest
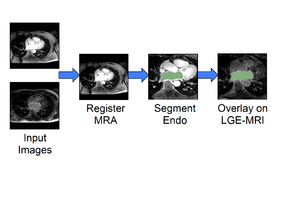
Cardiac Agatston Tutorial
|
CMR Toolkit LA workflow
|
Summer 2013 Tutorial contest
Cardiac MRI Toolkit
|
HelloCLI
|
SlicerRT
|
DTIPrep
|
Summer 2012 Tutorial contest
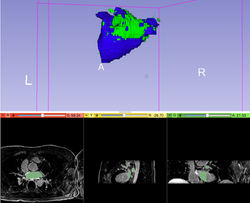
Automatic Left Atrial Scar Segmenter
|
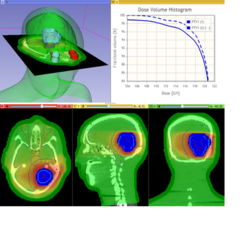
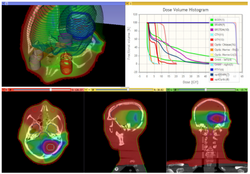
Qualitative and quantitative comparison of two RT dose distributions
|
Dose accumulation for adaptive radiation therapy
|
WebGL Export
|
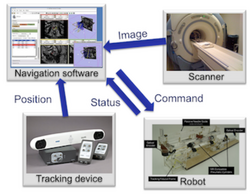
OpenIGTLink
|
Additional resources
|
|
|
|
|
|
External Resources
Using the Editor
This set of tutorials about the use of slicer in paleontology is very well written and provides step-by-step instructions. Even though it covers slicer version 3.4, many of the concepts and techniques have applicability to the new version and to any 3D imaging field:
- Open Source Paleontologist: 3D Slicer: The Tutorial
- Open Source Paleontologist: 3D Slicer: The Tutorial Part II
- Open Source Paleontologist: 3D Slicer: The Tutorial Part III
- Open Source Paleontologist: 3D Slicer: The Tutorial Part IV
- Open Source Paleontologist: 3D Slicer: The Tutorial Part V
- Open Source Paleontologist: 3D Slicer: The Tutorial Part VI
Team Contributions
See the collection of videos on the Kitware vimeo album.
User Contributions
See the User Contributions Page for more content.