Difference between revisions of "EMSegmenter-Tasks:CT-Hand-Bone"
Belhachemi (talk | contribs) |
Belhachemi (talk | contribs) |
||
| Line 47: | Line 47: | ||
* The atlas template scan and the subject scan are processed in the same way. | * The atlas template scan and the subject scan are processed in the same way. | ||
| + | |||
** Binary threshold image (based on Fiducials, TODO) | ** Binary threshold image (based on Fiducials, TODO) | ||
Extract the tissue and bones from the image. This could be done by specifying fiducials or by using fixed values(e.g. keep values between 150-3000 ) | Extract the tissue and bones from the image. This could be done by specifying fiducials or by using fixed values(e.g. keep values between 150-3000 ) | ||
| Line 56: | Line 57: | ||
** Binary image | ** Binary image | ||
| − | itk::BinaryThresholdImageFilter | + | Use itk::BinaryThresholdImageFilter to remove artifacts. Set everything between 0 and 30 to 0, otherwise 255 |
| − | |||
** extract largest component | ** extract largest component | ||
| − | itk::BinaryThresholdImageFilter | + | Use itk::BinaryThresholdImageFilter to extract label 255. |
| − | keep everything between 1 and 255 (1-254 are not used) and set it to one | + | <!-- keep everything between 1 and 255 (1-254 are not used) and set it to one --> |
Use itk::ConnectedComponentImageFilter to label the objects in the binary image. Each distinct object is assigned a unique label. | Use itk::ConnectedComponentImageFilter to label the objects in the binary image. Each distinct object is assigned a unique label. | ||
Revision as of 14:31, 25 April 2011
Home < EMSegmenter-Tasks:CT-Hand-BoneReturn to EMSegmenter Task Overview Page
Contents
Description
Single channel automatic segmentation of CT hand scans into the finger bones. The task can be applied to right and and left hand scans. The pipeline consist of the following steps:
- Step 1: Register the atlas to the CT scan via BRAINSFit (Johnson et al 2007)
- Step 1a:
- Step 1b:
- Step 1c:
- Step 2: Compute the intensity distributions for each structure
Compute intensity distribution (mean and variance) for each label by automatically sampling from the MR scan. The sampling for a specific label is constrained to the region that consists of voxels with high probability (top 95%) of being assigned to the label according to the aligned atlas.
- Step 4: Automatically segment the CT scan into the structures of interest using EM Algorithm (Pohl et al 2007)
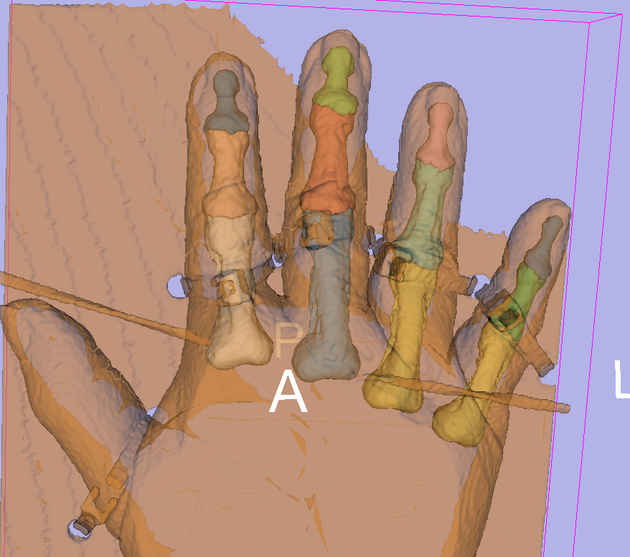
Anatomical Tree
The anatomical tree represents the structures to be segmented. Node labels displayed below contain a human readable structure name and in parentheses the internally used structure name.
- Hand
- Air
- Tissue
- Index finger / digitus secundus (II)
- Proximal (II)
- Medial (II)
- Distal (II)
- Middle finger / digitus medius (III)
- Proximal (III)
- Medial (III)
- Distal (III)
- Ring finger / digitus annularis (IV)
- Proximal (IV)
- Medial (IV)
- Distal (IV)
- Little finger / digitus minimus (V)
- Proximal (V)
- Medial (V)
- Distal (V)
Atlas
Pre-Processing
- Flip/Mirror the atlas (based on right hand scans) if left hand scan has to be segmented.
- The atlas template scan and the subject scan are processed in the same way.
- Binary threshold image (based on Fiducials, TODO)
Extract the tissue and bones from the image. This could be done by specifying fiducials or by using fixed values(e.g. keep values between 150-3000 )
- Blur image
itk::DiscreteGaussianImageFilter is used to fill remaining holes. (parameter e.g. Variance=1.5, MaximumKernelWidth 5)
- Binary image
Use itk::BinaryThresholdImageFilter to remove artifacts. Set everything between 0 and 30 to 0, otherwise 255
- extract largest component
Use itk::BinaryThresholdImageFilter to extract label 255.
Use itk::ConnectedComponentImageFilter to label the objects in the binary image. Each distinct object is assigned a unique label.
Use itk::RelabelComponentImageFilter to sort the labels based on the size of the object: the largest object will have label #1, the second largest will have label #2, etc.
Use itk::BinaryThresholdImageFilter to extract label 1. This is the largest object in the data set.
- Register the atlas template linear to the subject scan and save the linear transformation. (BRAINSFit Rigid,Affine)
- Register the atlas template non-linear to the subject scan. Use the linear transformation as initialization. (BRAINSDemonWarp)
Result
Collaborators
Vincent Magnotta (University of Iowa)
Acknowledgment
The construction of the pipeline was supported by funding from NIH NCRR 2P41RR013218 Supplement.
Citations
- Pohl K, Bouix S, Nakamura M, Rohlfing T, McCarley R, Kikinis R, Grimson W, Shenton M, Wells W. A Hierarchical Algorithm for MR Brain Image Parcellation. IEEE Transactions on Medical Imaging. 2007 Sept;26(9):1201-1212.
- S. Warfield, J. Rexilius, P. Huppi, T. Inder, E. Miller, W. Wells, G. Zientara, F. Jolesz, and R. Kikinis, “A binary entropy measure to assess nonrigid registration algorithms,” in MICCAI, LNCS, pp. 266–274, Springer, October 2001.
- Johnson H.J., Harris G., Williams K. BRAINSFit: Mutual Information Registrations of Whole-Brain 3D Images, Using the Insight Toolkit, The Insight Journal, July 2007
- T. Vercauteren, X. Pennec, A. Perchant, N. Ayache. Symmetric Log-Domain Diffeomorphic Registration: A Demons-based Approach. MICCAI 2008