Difference between revisions of "Slicer4:Diffusion"
m (→Mar 2011) |
m (→Mar 2011) |
||
| Line 115: | Line 115: | ||
** No correction will be done, if there are too many degenerate tensors, the user will have to use one of our noise/motion correction filters | ** No correction will be done, if there are too many degenerate tensors, the user will have to use one of our noise/motion correction filters | ||
** The mask generation will be separated from the tensor estimation | ** The mask generation will be separated from the tensor estimation | ||
| + | |||
| + | Deliverables Mar: CLI modules for tensor estimation and scalar extraction, tractography seeding. First version of DWI to Full brain tractography workflow | ||
==Apr 2011== | ==Apr 2011== | ||
Revision as of 19:57, 29 March 2011
Home < Slicer4:DiffusionBack to Slicer4 Projects
Contents
Introduction
This page contains descriptions and plans for the dMRI framework in Slicer 4
Periodical Meetings
Weekly Meeting
Every Tuesday from 3pm to 4pm at 1249 Boylston, 2nd floor conference room (LMI Suite)
Monthly Slicer4 coding sprint
Third Tuesday of every month, from 12pm to 4pm, Boylston 1249, 2nd floor conference room (bring your friends and significant programming others)
Upcoming Tuesdays:
- February 22nd (exceptional 4th Tuesday meeting)
- March 15th
Features to be addressed before the Summer NA-MIC meeting
- Porting/profiling of existing functionality (Alex)
- Getting CLI to a functioning state again (J2/JC)
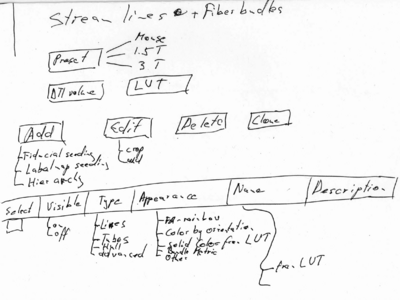
- Volumes module including DWI / Tensor
- Tensor estimation and scalar extraction
- Tractography seeding (fiducial/labelmap)
- Tensor Visualization (Basic visualization is priority)
- Tractography Visualization
- Displayable manager infrastructure
- Workflows (Demian)
- From DWI to full-brain tractography
- Peritumoral tractography
- DBP/High-end post-processing modules for tractography (Lauren)
- Cluster interaction
- Laterality analysis
- Tumor distance-transform (Isaiah)
- "Fluid" tractography exploration (Alex, this will be taken care of after the porting/profiling )
- Picking of fibers
- ROI-based selection of fibers
- Labelmap selection of fibers
- Usability testing / consultants (Isaiah, Ron, CF, Lauren, Demian)
- Extraction of statistical indices from a fiber bundle ( e.g.: Mean / std FA, length, ??volume?? )
- Test-cases for advanced functionality (Lauren/Antonio/Peter/Yogesh)
- Python support
- Multi-threading
- Weave (without SciPy)
- Fast LAPACK distribution (now comes with Slicer4)
- Matplotlib (maybe this will come with PySide)
- CLI Support (Jim Miller?)
- Diffusion CLI modules passing the data in memory as opposed to the actual file-based data interchange
- Python support
Schedule until June
Jan 2011
Kick-off, First meeting log
Define new infrastructure to allow efficient tractography (Demain, Lauren, Alex)
- Single ROI selection module
- Generalization to model hierarchies
- Labelmap selection module
- Bundle representation
- Reduce the memory footprint of bundles. First start, store in memory only the upper or lower triangular section of the matrix
- Port the fiducial tractography seeding to new infrastructure (Alex)
- Port tensor estimation tools (Demian)
- Implement quick and fluid exploration of pre-calculated full-brain tractographies
- Cluster interactive exploration, cluster labeling (Lauren)
- Fiber laterality module (Lauren)
- Define new infrastructure to allow several different representations of diffusivity (ODF, fODF, two-tensor, tensor, etc.)
Feb 2011
Feb 1, Discuss overall plan, distribution of responsibilities.
Feb 8, Review overall plan.
Feb 15, Definition of dependencies. Deadlines for top priority functionalities and some independent tools.
Feb 22, First code sprint.
- Assistants: Steve Pieper, Lauren O'Donnell, Antonio Tristán-Vega, Isaiah Norton and Demian Wassermann
- Main topics addressed:
- CLI interface for diffusion modules on Slicer 4 (and maybe 3): efficient ways of passing the volumes without having to dump them to disk all the time (Isaiah, Steve, Lauren, Demian and some help from Alex)
- Change of concept in slicer 4: from an application with a python interpreter embedded to a python application that can run inside of "any" given python interpreter (Steve, Lauren, Demian)
- Implementation of workflows within the newly developed ctkWorkflow framework (Demian)
- Finsler HARDI tractography (Antonio)
Deliverables Feb: Have detailed plan for March and April (Done!)
Mar 2011
Mar 1
- Getting CLI to a functioning state again (Demian is responsible - J2/JC will do the coding)
- Test the CLIs corresponding to (Isaiah)
- Tensor estimation
- Tensor Scalar extraction
- Tractography seeding (label-map)
Mar 8
Mar 15
- DWI to Full brain tractography workflow (Demian)
Mar 22
Mar 29
- Diffusion estimation issues with negative eigenvalues
- It was decided that the DT estimation will output a confidence image
- No correction will be done, if there are too many degenerate tensors, the user will have to use one of our noise/motion correction filters
- The mask generation will be separated from the tensor estimation
Deliverables Mar: CLI modules for tensor estimation and scalar extraction, tractography seeding. First version of DWI to Full brain tractography workflow
Apr 2011
Apr 5
- Finsler tractography CLI (Antonio)
- Basic Diffusion Volume Display: FA and color-by-orientation and Glyphs (Alex)
Apr 12
Apr 19
Apr 26
Deliverables Apr:
May 2011
May 3
May 10
May17
May 24
May 31
Deliverable May:
June 2011
June: debug, test
June (AHM),
Reference
Workflows
JJL & WJP tractography workflow/UI discussion
File format information
http://wiki.na-mic.org/Wiki/index.php/NAMIC_Wiki:Community_DTI