Difference between revisions of "Documentation/Nightly/Training"
Tag: 2017 source edit |
Tag: 2017 source edit |
||
| (26 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | =Introduction: Slicer Tutorials= | |
| − | |||
| − | =Introduction: Slicer | ||
| − | *This page contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment | + | *This page contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment to accomplish certain tasks. |
| − | + | *For "reference manual" style documentation, please see the [https://slicer.readthedocs.io/en/latest/ Slicer manual on ReadTheDocs]. | |
| − | *For "reference manual" style documentation, please | + | *For questions related to 3D Slicer training materials and to the organization of 3D Slicer training workshops, please send an e-mail to '''[https://scholar.harvard.edu/soniapujol/home Sonia Pujol, Ph.D., Director of Training and Education of 3D Slicer.]''' |
| − | *For questions related to the | ||
| − | + | *Some of these tutorials are based on older releases of 3D Slicer and are being upgraded to Slicer 5.0. The concepts are still useful but some interface elements and features may be different in updated versions. For tutorials for older versions of Slicer, please visit the [[Training| Slicer training portal]]. | |
| − | *Some of these tutorials are based on older releases of 3D Slicer and are being upgraded to | ||
__TOC__ | __TOC__ | ||
| Line 20: | Line 16: | ||
*The [https://spujol.github.io/3DSlicerQuickStartGuide/ Quick Start Guide] shows how to install and start 3D Slicer | *The [https://spujol.github.io/3DSlicerQuickStartGuide/ Quick Start Guide] shows how to install and start 3D Slicer | ||
*Author: Sonia Pujol, Ph.D. | *Author: Sonia Pujol, Ph.D. | ||
| − | *Based on | + | *Based on 3D Slicer 5.0 / 4.11 |
| align="right" | | | align="right" | | ||
[[image:QuickStart_image.png|250px|SlicerWelcome tutorial]] | [[image:QuickStart_image.png|250px|SlicerWelcome tutorial]] | ||
| Line 34: | Line 30: | ||
*Audience: First-time users who want a general introduction to the software | *Audience: First-time users who want a general introduction to the software | ||
*Modules: Welcome to Slicer, Sample Data | *Modules: Welcome to Slicer, Sample Data | ||
| − | *Based on | + | *Based on 3D Slicer 4.8 |
| + | *Compatible with Slicer 4.10.1 | ||
| align="right" | | | align="right" | | ||
[[image:SlicerWelcome-image.png|250px|SlicerWelcome tutorial]] | [[image:SlicerWelcome-image.png|250px|SlicerWelcome tutorial]] | ||
| Line 42: | Line 39: | ||
{| width="100%" | {| width="100%" | ||
| | | | ||
| − | *The [https://www.dropbox.com/s/ | + | *The [https://www.dropbox.com/s/v3lyivwgdoro7yn/Slicer4.10minute_SoniaPujol.pdf?dl=0| Slicer4 Minute Tutorial] is a brief introduction to the advanced 3D visualization capabilities of Slicer. |
*Author: Sonia Pujol, Ph.D. | *Author: Sonia Pujol, Ph.D. | ||
*Audience: First-time users who want to discover Slicer in 4 minutes | *Audience: First-time users who want to discover Slicer in 4 minutes | ||
*Modules: Welcome to Slicer, Models | *Modules: Welcome to Slicer, Models | ||
| − | *Based on | + | *Based on Slicer version 4.8 |
| + | *Compatible with Slicer 4.10.1 | ||
*The [[Media:Slicer4minute.zip|Slicer4Minute dataset]] contains an MR scan of the brain and 3D reconstructions of the anatomy | *The [[Media:Slicer4minute.zip|Slicer4Minute dataset]] contains an MR scan of the brain and 3D reconstructions of the anatomy | ||
| align="right" | | | align="right" | | ||
| Line 53: | Line 51: | ||
=3D Visualization= | =3D Visualization= | ||
| − | == | + | ==Data Loading and 3D Visualization== |
{| width="100%" | {| width="100%" | ||
| | | | ||
*Slicer 5.0 | *Slicer 5.0 | ||
| − | **The [https://spujol.github.io/SlicerVisualizationTutorial/ Slicer 5.0 Basics of data loading and visualization tutorial] shows how to load and visualize DICOM images and 3D models in 3D Slicer. | + | **The [https://spujol.github.io/SlicerVisualizationTutorial/ Slicer 5.0 Basics of data loading and visualization tutorial] shows how to load and visualize DICOM images and 3D models in 3D Slicer. [https://docs.google.com/presentation/d/12Lbq-QBCxP2p9FkF3_YM5Ng7pItfspMG0FP_20wQglA/edit?usp=sharing French version] |
**Author: Sonia Pujol, Ph.D. | **Author: Sonia Pujol, Ph.D. | ||
**Modules: DICOM, Volume Rendering, Models | **Modules: DICOM, Volume Rendering, Models | ||
| Line 63: | Line 61: | ||
**Based on: 3D Slicer version 5.0/4.11 | **Based on: 3D Slicer version 5.0/4.11 | ||
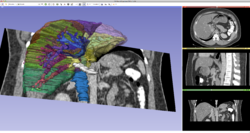
**The [https://www.dropbox.com/s/03emcqnlec4t2s5/3DVisualizationDataset.zip?dl=1 Data Loading and Visualization dataset] contains a thoraco-abdominal CT scan, an MRI brain dataset and 3D models of brain structures. | **The [https://www.dropbox.com/s/03emcqnlec4t2s5/3DVisualizationDataset.zip?dl=1 Data Loading and Visualization dataset] contains a thoraco-abdominal CT scan, an MRI brain dataset and 3D models of brain structures. | ||
| − | |||
| Line 71: | Line 68: | ||
**Modules: Welcome to Slicer, Data, Volume Rendering, Models. | **Modules: Welcome to Slicer, Data, Volume Rendering, Models. | ||
**Audience: End-users | **Audience: End-users | ||
| − | **Based on | + | **Based on Slicer 4.9 |
| − | **The [http://slicer.kitware.com/midas3/download/?items=330421,1 3DVisualization dataset] | + | **Compatible with Slicer 4.10.1 |
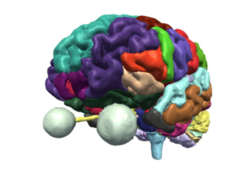
| + | **The [http://slicer.kitware.com/midas3/download/?items=330421,1 3DVisualization dataset] contains an MR scan and a series of 3D models of the brain. | ||
| align="right" | | | align="right" | | ||
| Line 79: | Line 77: | ||
|} | |} | ||
| − | == | + | ==DICOM== |
| + | {| width="100%" | ||
| + | | | ||
| + | *The [https://spujol.github.io/SlicerDICOMTutorial/ DICOM and Slicer] tutorial provides an introduction to the DICOM standard and shows how to load and visualize DICOM datasets in 3D Slicer version 5.0. | ||
| + | *Author: Sonia Pujol, Ph.D. | ||
| + | *Modules: DICOM, Volumes | ||
| + | *Based on: 3D Slicer version 5.0/4.11 | ||
| + | *The [https://spujol.github.io/SlicerDICOMTutorial/ 3D Slicer DICOM Tutorial Data] contains a torso-CT and a breast MRI. | ||
| + | | align="right" | | ||
| + | [[Image:SlicerAndDICOM.png|right|250px|]] | ||
| + | |} | ||
| + | |||
{| width="100%" | {| width="100%" | ||
| | | | ||
| − | *The [https://www.dropbox.com/s/ | + | *The [https://www.dropbox.com/s/8pm5mty2c0zwmyk/3DVisualizationDICOM_Slicer4.10_SoniaPujol.pdf?dl=0 3D Visualization of DICOM images] course guides through 3D data loading and visualization of DICOM images for Radiology Applications in Slicer4. |
*Author: Sonia Pujol, Ph.D., Kitt Shaffer, M.D., Ph.D., Ron Kikinis, M.D. | *Author: Sonia Pujol, Ph.D., Kitt Shaffer, M.D., Ph.D., Ron Kikinis, M.D. | ||
*Audience: Radiologists and users of Slicer who need a more comprehensive overview over Slicer4 visualization capabilities. | *Audience: Radiologists and users of Slicer who need a more comprehensive overview over Slicer4 visualization capabilities. | ||
*Modules: DICOM, Volumes, Volume Rendering, Models. | *Modules: DICOM, Volumes, Volume Rendering, Models. | ||
*Based on: 3D Slicer version 4.8 | *Based on: 3D Slicer version 4.8 | ||
| + | *Compatible with 3D Slicer version 4.10 | ||
*The [[Media:3DVisualization DICOM images part1.zip| 3DVisualizationDICOM_part1]] and [[Media:3DVisualization DICOM images part2.zip| 3DVisualizationDICOM_part2]] datasets contain a series of MR and CT scans, and 3D models of the brain, lung and liver. | *The [[Media:3DVisualization DICOM images part1.zip| 3DVisualizationDICOM_part1]] and [[Media:3DVisualization DICOM images part2.zip| 3DVisualizationDICOM_part2]] datasets contain a series of MR and CT scans, and 3D models of the brain, lung and liver. | ||
| align="right" | | | align="right" | | ||
| Line 92: | Line 102: | ||
|} | |} | ||
| + | ==Open Anatomy Browser== | ||
| + | {| width="100%" | ||
| + | | | ||
| + | [[Image:OABrowser.png|right|250px|]] | ||
| − | = | + | *The [https://www.dropbox.com/s/f2641iu27hif8p4/OpenAnatomyTutorial_SoniaPujol-MikeHalle.pdf?dl=0 Open Anatomy Browser] tutorial is an introduction to the OABrowser technology for viewing and interacting with atlases. |
| + | *Author: Sonia Pujol, Ph.D., Mike Halle, Ph.D. | ||
| + | *Audience: End-users | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
|} | |} | ||
| + | |||
| + | =Tutorials for software developers= | ||
==PerkLab's Slicer bootcamp training materials== | ==PerkLab's Slicer bootcamp training materials== | ||
| Line 117: | Line 126: | ||
| align="right" | | | align="right" | | ||
[[Image:PerkLabSlicerProgrammingTutorial.png|right|250px|]] | [[Image:PerkLabSlicerProgrammingTutorial.png|right|250px|]] | ||
| + | |} | ||
| + | |||
| + | ==Slicer Programming Tutorial== | ||
| + | {| width="100%" | ||
| + | | | ||
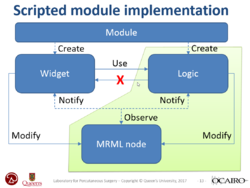
| + | *The [https://spujol.github.io/SlicerProgrammingTutorial/ Slicer Programming tutorial] guides through the integration of a python module in Slicer. It provides an introduction to the Python Console and the Qt Widget toolkit in 3D Slicer. | ||
| + | *Author: Sonia Pujol, Ph.D., Steve Pieper, Ph.D. | ||
| + | *Audience: Developers | ||
| + | *Based on: 3D Slicer version 5.0/4.11 | ||
| + | | align="right" | | ||
| + | [[Image:SlicerProgrammingTutorial.png|right|250px|]] | ||
|} | |} | ||
==Slicer script repository== | ==Slicer script repository== | ||
| − | For additional Python scripts examples, please visit the [ | + | For additional Python scripts examples, please visit the [https://slicer.readthedocs.io/en/latest/developer_guide/script_repository.html Script repository]. |
==Developing and contributing extensions for 3D Slicer== | ==Developing and contributing extensions for 3D Slicer== | ||
| Line 135: | Line 155: | ||
=Segmentation= | =Segmentation= | ||
| − | |||
| − | |||
{| width="100%" | {| width="100%" | ||
| | | | ||
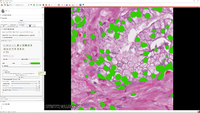
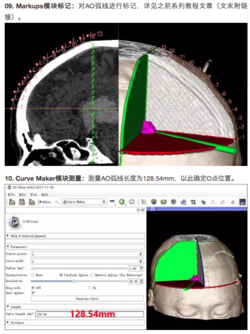
*Segmentation for 3D printing: shows how to use the Segment Editor module for combining CAD designed parts with patient-specific models. | *Segmentation for 3D printing: shows how to use the Segment Editor module for combining CAD designed parts with patient-specific models. | ||
**'''[https://discourse.slicer.org/t/new-video-tutorial-for-segment-editor-lumbar-spine-segmentation-for-3d-printing/700 Video tutorial]'''. Author: Hillary Lia. | **'''[https://discourse.slicer.org/t/new-video-tutorial-for-segment-editor-lumbar-spine-segmentation-for-3d-printing/700 Video tutorial]'''. Author: Hillary Lia. | ||
| − | **'''[[Documentation/{{documentation/version}}/Training#Segmentation_for_3D_printing|Segmentation for 3D printing Step-by-step tutorial]]'''. Author: Csaba Pinter | + | **'''[[Documentation/{{documentation/version}}/Training#Segmentation_for_3D_printing|Segmentation for 3D printing Step-by-step tutorial]]'''. Author: Csaba Pinter, MSc |
**Audience: Users and developers interested in segmentation and 3D printing | **Audience: Users and developers interested in segmentation and 3D printing | ||
**Dataset: [[:File:BasePiece.zip|Phantom base STL model]] Source: [http://perk-software.cs.queensu.ca/plus/doc/nightly/modelcatalog/ PerkLab]. | **Dataset: [[:File:BasePiece.zip|Phantom base STL model]] Source: [http://perk-software.cs.queensu.ca/plus/doc/nightly/modelcatalog/ PerkLab]. | ||
| Line 148: | Line 166: | ||
|--- | |--- | ||
| | | | ||
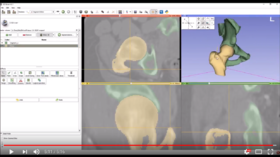
| − | *'''[https:// | + | *'''[https://www.youtube.com/watch?v=BJoIexIvtGo Video tutorial: Whole heart segmentation from cardiac CT]''' shows how to use the Segment Editor module for segmenting heart ventricles, atria, and great vessels from cardiac CT volumes. |
**Author: Andras Lasso, PhD | **Author: Andras Lasso, PhD | ||
**Audience: Users who need to segment heart structures, for example for visualization, quantification, or simulation. | **Audience: Users who need to segment heart structures, for example for visualization, quantification, or simulation. | ||
| − | ** | + | **[http://slicer.kitware.com/midas3/download/bitstream/738905/CTA-cardio2.nrrd Sample data set] |
**Based on: 3D Slicer version 4.8 | **Based on: 3D Slicer version 4.8 | ||
| align="right" |[[Image:WholeHeartSegYoutube.png|280px]] | | align="right" |[[Image:WholeHeartSegYoutube.png|280px]] | ||
|--- | |--- | ||
| | | | ||
| − | *'''[https:// | + | *'''[https://www.youtube.com/watch?v=0at15gjk-Ns Video tutorial: Femur and pelvis segmentation from CT]''' shows how to use the Segment Editor module for segmenting pelvis and femur from CT volumes. |
**Author: Andras Lasso, PhD | **Author: Andras Lasso, PhD | ||
**Audience: Users who need to segment bones in CT images for visualization, quantification, or simulation. | **Audience: Users who need to segment bones in CT images for visualization, quantification, or simulation. | ||
| Line 164: | Line 182: | ||
|--- | |--- | ||
| | | | ||
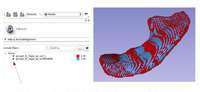
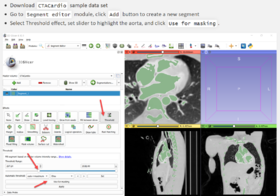
| − | *[https://lassoan.github.io/SlicerSegmentationRecipes/ Slicer Segmentation Recipes] | + | *'''[https://lassoan.github.io/SlicerSegmentationRecipes/ Slicer Segmentation Recipes]''' provide step-by-step description of useful segmentation techniques. |
| + | ** Segmentation tutorials for common tasks, such as skin surface extraction, craniotomy (splitting segments), sorta segmentation, cerebral vessel segmentation by subtraction, segmentation on arbitrarily oriented slices, skull stripping. | ||
| + | | align="right" |[[Image:SegmentationRecipes.png|280px]] | ||
|--- | |--- | ||
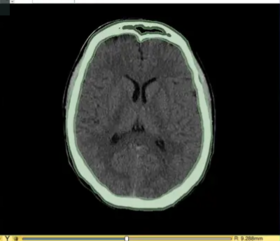
| | | | ||
| Line 172: | Line 192: | ||
**Based on: 3D Slicer version 4.11 | **Based on: 3D Slicer version 4.11 | ||
| align="right" |[[Image:SkullStripping.png|280px]] | | align="right" |[[Image:SkullStripping.png|280px]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | =Image Phenotyping= | ||
| + | {| width="100%" | ||
| + | | | ||
| + | *Based on: 3D Slicer version 4.10 | ||
| + | *The [https://spujol.github.io/ImagePhenotypingTutorial/ Image Phenotyping tutorial] is an introduction to brain tumor segmentation and image phenotyping using the Slicer Radiomics extension. | ||
| + | *Authors: Sonia Pujol, Ph.D. | ||
| + | *Audience: Clinical researchers | ||
| + | *Dataset: [https://www.dropbox.com/s/hdlduw6oqnf2n72/Meningioma.nrrd?dl=0 Meningioma dataset] | ||
| + | | align="right" |[[File:ImagePhenotyping.png|250px]] | ||
| + | |||
|} | |} | ||
| Line 187: | Line 220: | ||
|} | |} | ||
| − | *Based on: 3D Slicer version 4.8 | + | *Based on: 3D Slicer version 4.8; Compatible with Slicer 4.10 |
| + | |||
| + | {| width="100%" | ||
| + | | | ||
| + | *The [https://spujol.github.io/3DSlicerTutorial-Registration Brain Tumor Registration] is a video-based tutorial that shows how to register two MRI datasets in a brain tumor case for surgical resection follow-up. | ||
| + | *Authors: Sonia Pujol, Ph.D., Dominik Meier, Ph.D. | ||
| + | *Audience: Users and developers interested in image registration | ||
| + | *Dataset: [[Special:FilePath/RegLib C37 Data.zip| Registration Library Case #37]] | ||
| + | | align="right" |[[File:RigidRegistration.jpg|250px]] | ||
| + | |} | ||
| + | *Based on: 3D Slicer version 4.10 | ||
==Slicer Registration Case Library== | ==Slicer Registration Case Library== | ||
| − | |||
{| width="100%" | {| width="100%" | ||
| | | | ||
| Line 198: | Line 240: | ||
:Author: Dominik Meier, Ph.D. | :Author: Dominik Meier, Ph.D. | ||
:Audience: users interested learning/applying Slicer image registration technology | :Audience: users interested learning/applying Slicer image registration technology | ||
| − | | align="right" |[[Image:RegLib_table.png|250px|link=https://www.slicer.org/wiki/Documentation/{{documentation/version}}/Registration/RegistrationLibrary]] | + | | align="right" |[[Image:RegLib_table.png|250px|link=https://www.slicer.org/wiki/Documentation/{{documentation/version}}/Registration/RegistrationLibrary]] |
|} | |} | ||
| − | + | ||
| + | =Slicer Extensions= | ||
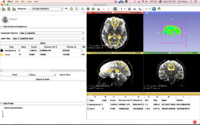
==Slicer4 Diffusion Tensor Imaging Tutorial== | ==Slicer4 Diffusion Tensor Imaging Tutorial== | ||
{| width="100%" | {| width="100%" | ||
| | | | ||
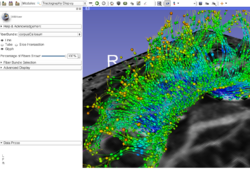
| − | + | *The [https://spujol.github.io/SlicerDiffusionMRITutorial Diffusion MRI Tutorial] is an introduction to the basics of loading diffusion weighted images in Slicer, estimating tensors and generating fiber tracts. | |
| − | *The [https:// | ||
*Author: Sonia Pujol, Ph.D. | *Author: Sonia Pujol, Ph.D. | ||
*Audience: End-users and developers | *Audience: End-users and developers | ||
| − | *Modules: Data, Volumes, DWI to DTI Estimation, Diffusion Tensor Scalar Measurements, Editor, Markups,Tractography Label Map Seeding, Tractography Interactive Seeding | + | *Modules: Data, Volumes, DWI to DTI Estimation, Diffusion Tensor Scalar Measurements, Editor, Markups, Tractography Label Map Seeding, Tractography Interactive Seeding |
| − | *Based on: 3D Slicer version 4.8 | + | *Based on: 3D Slicer version 4.8; Compatible with Slicer version 4.10.2 |
| − | *The [ | + | *The [https://www.dropbox.com/s/gba2zsn276x43up/SlicerDiffusionMRITutorialData.zip?dl=1 Slicer Diffusion MRI Tutorial dataset] contains an MR Diffusion Weighted Imaging scan of the brain. |
| + | *Please visit [http://dmri.slicer.org/docs/ dmri.slicer.org/docs] for the latest documentation of SlicerDMRI. | ||
| align="right" | | | align="right" | | ||
[[Image:Slicer4DTI Tutorial.png|right|250px|]] | [[Image:Slicer4DTI Tutorial.png|right|250px|]] | ||
| Line 295: | Line 338: | ||
| align="right" | | | align="right" | | ||
[[File:SlicerWinterProjectWeek2017-FiberBundleVolumeMeasurements.png | 200px]]. | [[File:SlicerWinterProjectWeek2017-FiberBundleVolumeMeasurements.png | 200px]]. | ||
| + | |} | ||
| + | |||
| + | ==Slicer4 Lung CT Analyzer== | ||
| + | {| width="100%" | ||
| + | | | ||
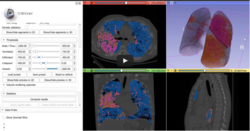
| + | *The [https://github.com/rbumm/SlicerLungCTAnalyzer LungCTAnalyzer tutorial] and the [https://www.youtube.com/watch?v=fpLxm7uAvZQ LungCTAnalyzer video-based demo] show how to visualize and quantify infiltration, emphysema and collapsed lung areas in CT datasets acquired on COVID-19 patients. | ||
| + | *Authors: Rudolph Bumm, MD, Andras Lasso, PhD. | ||
| + | *Audience: End-users | ||
| + | *Modules: LungCTSegmenter, LungCTAnalyzer | ||
| + | *Based on: 3D Slicer version 5.0 (4.11) | ||
| + | | align="right" | | ||
| + | [[Image:LungCTAnalyzer.png|right|250px|]] | ||
| + | |||
|} | |} | ||
| Line 368: | Line 424: | ||
| style="width:33%" |[[Image:3DPrinting.png|right|250px|]] [https://www.youtube.com/watch?v=MKLWzD0PiIc Preparing data for 3D printing - Author: Nabgha Farhat] | | style="width:33%" |[[Image:3DPrinting.png|right|250px|]] [https://www.youtube.com/watch?v=MKLWzD0PiIc Preparing data for 3D printing - Author: Nabgha Farhat] | ||
|} | |} | ||
| − | {| border="1" cellpadding="5" width="1200px" | + | {| border="1" cellpadding="5" width="1200px" | |
| − | | style="width: | + | | style="width:25%" |[[Image:DICOM2.png|right|250px|]] [https://www.youtube.com/watch?v=nzWf4xHy1BM& How to export CT and segmentation data to DICOM- Author: Andras Lasso, Csaba Pinter] |
| − | | style="width: | + | | style="width:25%" |[[Image:LocalThresholdEffect.png|right|250px|]] [https://www.youtube.com/watch?time_continue=26&v=cevlMLyhfK8&feature=emb_logo Local Threshold Effect - Author: Kyle Sunderland] |
| − | | style="width: | + | | style="width:25%" |[[Image:VMTKCenterlines.png|right|250px|]] [https://www.youtube.com/watch?v=yi07mjr3JeU SlicerVMTK centerline extraction (Slicer 4.11)- Author: Andras Lasso] |
| − | |- | + | | style="width:25%" |[[Image:MONAILabel.png|right|250px|]] [https://www.youtube.com/watch?v=PmD8umlcpF4 MONAI Label(Slicer 4.11)- Author: Andres Diaz-Pinto] |
|} | |} | ||
| Line 419: | Line 475: | ||
*[https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247486025&idx=1&sn=b281324893be4ab116d20826f1b426c3&chksm=eacc007bddbb896d9deb096f209278f40c0b52c6410a8a9ff3ce8c3697c99304f18eb678f11e&mpshare=1&scene=24&srcid=02125v1kxvIGmfkxx7mUZcCM#rd Mobile phone positioning and AR application 手机定位及AR应用的初步探索] | *[https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247486025&idx=1&sn=b281324893be4ab116d20826f1b426c3&chksm=eacc007bddbb896d9deb096f209278f40c0b52c6410a8a9ff3ce8c3697c99304f18eb678f11e&mpshare=1&scene=24&srcid=02125v1kxvIGmfkxx7mUZcCM#rd Mobile phone positioning and AR application 手机定位及AR应用的初步探索] | ||
| + | *[https://mp.weixin.qq.com/s?__biz=MzI3MDY4ODA5Mw==&mid=2247483658&idx=1&sn=ad08fe01c61d6999a36f2960b34287ec&chksm=eacc0b38ddbb822e60206afcf0bb67562432bb275463b20ad6ac7d243ccc1429afaa8f2177ea#rd 3D printing 如何用3D Slicer实现模型3D打印 束旭俊] | ||
| + | |||
| + | The WeChat 3D Slicer Group in China offers a [https://spujol.github.io/SlicerTutorialsInChinese/ comprehensive list of tutorials in Chinese.] | ||
| + | <br /> | ||
| − | |||
| align="right" | | | align="right" | | ||
[[image:Wechat-hemorage-2018-02-12.png|250px|Example WeChat tutorial slides]] | [[image:Wechat-hemorage-2018-02-12.png|250px|Example WeChat tutorial slides]] | ||
| Line 447: | Line 506: | ||
*Audience: Users interested in segmentation | *Audience: Users interested in segmentation | ||
| align="right" |[[File:FastGrowCutLogo.png|200px]] | | align="right" |[[File:FastGrowCutLogo.png|200px]] | ||
| + | |} | ||
| + | |||
| + | {| width="100%" | ||
| + | *'''[https://www.youtube.com/channel/UC8vxI0-dEWrw0_tBF-v8xGA/videos Video-based segmentation tutorials from CHU de Rouen (France)] | ||
| + | ** Segmentation tutorials, including liver, wrist bones, lungs, kidneys, hips. | ||
| + | | align="right" |[[Image:ChuRouen.png|180px]] | ||
|} | |} | ||
| Line 459: | Line 524: | ||
*[http://openpaleo.blogspot.com/2009/03/3d-slicer-tutorial-part-v.html Open Source Paleontologist: 3D Slicer: The Tutorial Part V] | *[http://openpaleo.blogspot.com/2009/03/3d-slicer-tutorial-part-v.html Open Source Paleontologist: 3D Slicer: The Tutorial Part V] | ||
*[http://openpaleo.blogspot.com/2009/03/3d-slicer-tutorial-part-vi.html Open Source Paleontologist: 3D Slicer: The Tutorial Part VI] | *[http://openpaleo.blogspot.com/2009/03/3d-slicer-tutorial-part-vi.html Open Source Paleontologist: 3D Slicer: The Tutorial Part VI] | ||
| − | |||
| − | |||
Latest revision as of 23:40, 22 November 2022
Home < Documentation < Nightly < TrainingIntroduction: Slicer Tutorials
- This page contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment to accomplish certain tasks.
- For "reference manual" style documentation, please see the Slicer manual on ReadTheDocs.
- For questions related to 3D Slicer training materials and to the organization of 3D Slicer training workshops, please send an e-mail to Sonia Pujol, Ph.D., Director of Training and Education of 3D Slicer.
- Some of these tutorials are based on older releases of 3D Slicer and are being upgraded to Slicer 5.0. The concepts are still useful but some interface elements and features may be different in updated versions. For tutorials for older versions of Slicer, please visit the Slicer training portal.
Contents
Quick Start Guide
Downloading and Installing Slicer
|
General Introduction
Slicer Welcome Tutorial
|
Slicer4Minute Tutorial
|
3D Visualization
Data Loading and 3D Visualization
|
DICOM
|
|
Open Anatomy Browser
|
Tutorials for software developers
PerkLab's Slicer bootcamp training materials
|
Slicer Programming Tutorial
|
Slicer script repository
For additional Python scripts examples, please visit the Script repository.
Developing and contributing extensions for 3D Slicer
|
Segmentation
|
|
|
|
|
|
|
|
|
|
Image Phenotyping
|
|
Registration
Slicer4 Image Registration
|
|
- Based on: 3D Slicer version 4.8; Compatible with Slicer 4.10
|
|
- Based on: 3D Slicer version 4.10
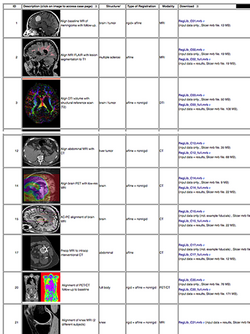
Slicer Registration Case Library
|
|
Slicer Extensions
Slicer4 Diffusion Tensor Imaging Tutorial
|
Slicer4 Neurosurgical Planning Tutorial
|
Slicer4 Quantitative Imaging tutorial
|
Slicer4 IGT
|
Slicer4 Radiation Therapy Tutorial
|
Slicer Pathology
|
SPHARM-PDM
|
Fiber Bundle Volume Measurement
|
Slicer4 Lung CT Analyzer
|
3D Slicer version 4.7 Tutorial Contest
For previous editions of the contest, please visit the 3D Slicer Tutorial Contests page
Segmentation for 3D printing
|
Slicer Pathology
|
Simple Python Tool for Quality Control of DWI data
|
SPHARM-PDM
|
Integration of Robot Operating System (ROS) and 3D Slicer using OpenIGTLink
|
Fiber Bundle Volume Measurement
|
YouTube videos
Additional non-curated videos-based demonstrations using 3D Slicer are accessible on YouTube.
Teams Contributions
|
|
|
|
International resources
International resources in Chinese and in German are made available by the Slicer community.
Resources in Chinese
|
A 3D Slicer community on WeChat in China offers many tutorials and clinical examples in Chinese. Note that the images are of interest to non-Chinese speakers and Google Translate does a reasonable job of translating some of the text. The tutorials below are examples of Slicer tutorials in Chinese. The WeChat 3D Slicer Group in China offers a comprehensive list of tutorials in Chinese.
|
Resources in German
- A series of four YouTube videos on python programming in Slicer (German narration with English subtitles)
Murat Maga's blog posts about using 3D Slicer for biology
Using the (legacy) Editor
Fast GrowCut
|

|
- Video-based segmentation tutorials from CHU de Rouen (France)
- Segmentation tutorials, including liver, wrist bones, lungs, kidneys, hips.
|
Use case: Slicer in paleontology
This set of tutorials about the use of slicer in paleontology is very well written and provides step-by-step instructions. Even though it covers slicer version 3.4, many of the concepts and techniques have applicability to the new version and to any 3D imaging field:
- Open Source Paleontologist: 3D Slicer: The Tutorial
- Open Source Paleontologist: 3D Slicer: The Tutorial Part II
- Open Source Paleontologist: 3D Slicer: The Tutorial Part III
- Open Source Paleontologist: 3D Slicer: The Tutorial Part IV
- Open Source Paleontologist: 3D Slicer: The Tutorial Part V
- Open Source Paleontologist: 3D Slicer: The Tutorial Part VI