Difference between revisions of "Documentation/Nightly/Modules/GeodesicSlicer"
(add DOI) Tag: 2017 source edit |
|||
| (14 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | <noinclude>{{documentation/versioncheck}}</noinclude> | + | <noinclude>{{documentation/versioncheck}} |
| + | </noinclude> | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-header}} | {{documentation/{{documentation/version}}/module-header}} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| − | [[File:GeodesicSlicer logo.png|128x128px|thumb|left| | + | [[File:GeodesicSlicer logo.png|128x128px|thumb|left|]] |
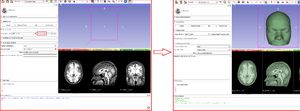
[[File:Screen-shot of the GeodesicSlicer program. .png|thumb|right|The users enters 1) the T1-weighted whole-brain anatomical image 2) Place four points: the nasion, the inion, the left tragus and the right tragus. The program make a 3D mesh morphed to the structural MRI data of a participant and calculates the 10-20 system EEG with T3P3, and outputs the distance between the anatomical target and the T3 electrode.]] | [[File:Screen-shot of the GeodesicSlicer program. .png|thumb|right|The users enters 1) the T1-weighted whole-brain anatomical image 2) Place four points: the nasion, the inion, the left tragus and the right tragus. The program make a 3D mesh morphed to the structural MRI data of a participant and calculates the 10-20 system EEG with T3P3, and outputs the distance between the anatomical target and the T3 electrode.]] | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 14: | Line 15: | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
{{documentation/{{documentation/version}}/module-introduction-end}} | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
| + | |||
The module has been developed based on ideas and feedbacks from the community. We would like to especially thank: | The module has been developed based on ideas and feedbacks from the community. We would like to especially thank: | ||
| − | * Dr. Olivier Etard, M.D., Ph.D., CHU de Caen. | + | *Dr. Olivier Etard, M.D., Ph.D., CHU de Caen. |
| − | * Dr. Clément Nathou, M.D., Ph.D., CHU de Caen. | + | *Dr. Clément Nathou, M.D., Ph.D., CHU de Caen. |
| − | * Dr. Nicolas Delcroix, Ph.D., UMS 3408. | + | *Dr. Nicolas Delcroix, Ph.D., UMS 3408. |
| − | * Dr. Sonia Dollfus, M.D., Ph.D., CHU de Caen, header of [http://www.ists.cyceron.fr/spip.php?rubrique17 ISTS]. | + | *Dr. Sonia Dollfus, M.D., Ph.D., CHU de Caen, header of [http://www.ists.cyceron.fr/spip.php?rubrique17 ISTS]. |
| − | * Dr. Csaba Pinter, MSc, Queen's University. | + | *Dr. Csaba Pinter, MSc, Queen's University. |
| − | * Dr. Andras Lasso, Ph.D., Queen's University. | + | *Dr. Andras Lasso, Ph.D., Queen's University. |
| − | ''If you use this module, please cite the following article: <ref name="Briend | + | ''If you use this module, please cite the following article: <ref name="Briend 2020a">Briend F. et al., A new toolbox to compare NIBS localization method: Application for auditory hallucinations in schizophrenia. Schizophrenia Research, https://doi.org/10.1016/j.schres.2020.09.001</ref> and <ref name="Briend 2020b">Briend F. et al., GeodesicSlicer: a Slicer Toolbox for Targeting Brain Stimulation. Neuroinformatics. https://doi.org/10.1007/s12021-020-09457-9</ref>.'' |
{{Warning |This extension is under the [http://www.cecill.info/licences/Licence_CeCILL_V2.1-en.html CeCill license], a copyleft license.}} | {{Warning |This extension is under the [http://www.cecill.info/licences/Licence_CeCILL_V2.1-en.html CeCill license], a copyleft license.}} | ||
| + | |||
__TOC__ | __TOC__ | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 33: | Line 36: | ||
'''Terminology''' | '''Terminology''' | ||
| + | |||
*'''''Mesh''''' A mesh or polygon mesh is a collection of vertices, edges and faces that defines the shape of a polyhedral object in 3D computer graphics and solid modeling. | *'''''Mesh''''' A mesh or polygon mesh is a collection of vertices, edges and faces that defines the shape of a polyhedral object in 3D computer graphics and solid modeling. | ||
*'''''Shortest path''''' In graph theory, the shortest path problem is the problem of finding a path between two vertices (or nodes) in a graph such that the sum of the weights of its constituent edges is minimized. | *'''''Shortest path''''' In graph theory, the shortest path problem is the problem of finding a path between two vertices (or nodes) in a graph such that the sum of the weights of its constituent edges is minimized. | ||
| − | *'''''10-20 EEG system''''' The International 10-20 system is commonly used for electroencephalogram (EEG) electrode placement and to correlate external skull locations with underlying cortical areas.<ref name="Jasper 1958">Jasper, H. (1958). The ten twenty electrode system of the international federation. Electroencephalography and Clinical Neurophysiology, 10, 371‑375.</ref> | + | *'''''10-20 EEG system''''' The International 10-20 system is commonly used for electroencephalogram (EEG) electrode placement and to correlate external skull locations with underlying cortical areas.<ref name="Jasper 1958">Jasper, H. (1958). The ten twenty electrode system of the international federation. Electroencephalography and Clinical Neurophysiology, 10, 371‑375.</ref> |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| − | {{documentation/{{documentation/version}}/module-section|Installation | + | {{documentation/{{documentation/version}}/module-section|Installation}} |
| − | # First, open 3D Slicer | + | |
| − | # Open the Slicer Extensions from the icon on the menu bar | + | #First, open 3D Slicer |
| − | # Choose "Geodesic Slicer" module from the list of extensions and click "INSTALL" button. | + | #Open the Slicer Extensions from the icon on the menu bar |
| − | # Once you restart 3D Slicer, the Geodesic Slicer module should show up on the Modules menu (under Informatics->Geodesic Slicer) | + | #Choose "Geodesic Slicer" module from the list of extensions and click "INSTALL" button. |
| + | #Once you restart 3D Slicer, the Geodesic Slicer module should show up on the Modules menu (under Informatics->Geodesic Slicer) | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Use Cases}} | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
The overall goal is to allow users to find the shortest paths between nodes in a graph and via the Dijkstra's algorithm to make 10-20 system. This module can be used for: | The overall goal is to allow users to find the shortest paths between nodes in a graph and via the Dijkstra's algorithm to make 10-20 system. This module can be used for: | ||
| + | |||
*Stimulation in psychiatry: MRI guided brain stimulation without the use of a neuronavigation system. | *Stimulation in psychiatry: MRI guided brain stimulation without the use of a neuronavigation system. | ||
*Surgery measurement. | *Surgery measurement. | ||
| Line 54: | Line 60: | ||
{{documentation/{{documentation/version}}/module-section|Panels and their use}} | {{documentation/{{documentation/version}}/module-section|Panels and their use}} | ||
| − | ==== Create a mesh ==== | + | ====Create a mesh==== |
[[File:Create mesh.png|thumb|right]] | [[File:Create mesh.png|thumb|right]] | ||
A typical straightforward Geodesic Slicer workflow for consists of the following steps: | A typical straightforward Geodesic Slicer workflow for consists of the following steps: | ||
| − | # Load a volume.nii (by Drag & Drop or the Add Data dialogue). | + | #Load a volume.nii (by Drag & Drop or the Add Data dialogue). |
| − | # Enter in the Geodesic Slicer module using either the toolbar or the Modules menu button. | + | #Enter in the Geodesic Slicer module using either the toolbar or the Modules menu button. |
| − | # Press the button "Create a quick mesh" or "Create a mesh" (with filling holes smoothing, better for the next part but longer). | + | #Press the button "Create a quick mesh" or "Create a mesh" (with filling holes smoothing, better for the next part but longer). |
#*Wait a moment. | #*Wait a moment. | ||
#Go to '''Parameters to find the shortest path''' or '''Make 10-20 EEG system electrode''' section. | #Go to '''Parameters to find the shortest path''' or '''Make 10-20 EEG system electrode''' section. | ||
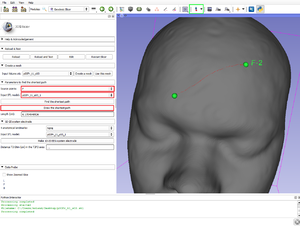
| − | ==== Parameters to find the shortest path ==== | + | ====Parameters to find the shortest path==== |
[[File:Shortest past.png|thumb|right]] | [[File:Shortest past.png|thumb|right]] | ||
| − | # Source points: The list of fiducial points on the curve, since the "Create-and-place Fiducial" button (in green in the figure above). | + | #Source points: The list of fiducial points on the curve, since the "Create-and-place Fiducial" button (in green in the figure above). |
| − | # Input STL model: The model you use (after "use this mesh", the T1.stl created). | + | #Input STL model: The model you use (after "use this mesh", the T1.stl created). |
| − | #* Find the shortest path: Calculate in centimeter the geodesic (shortest) path via the Dijkstra's algorithm. | + | #*Find the shortest path: Calculate in centimeter the geodesic (shortest) path via the Dijkstra's algorithm. |
| − | #* Draw the shortest path: Draw the Dijkstra's algorithm shortest path. | + | #*Draw the shortest path: Draw the Dijkstra's algorithm shortest path. |
| − | #** Length (cm): The length of the current curve is shown in centimeter. | + | #**Length (cm): The length of the current curve is shown in centimeter. |
====10-20 system electrode==== | ====10-20 system electrode==== | ||
| + | |||
*Run the Dijkstra's algorithm to '''make the 10-20 system electrode'''. | *Run the Dijkstra's algorithm to '''make the 10-20 system electrode'''. | ||
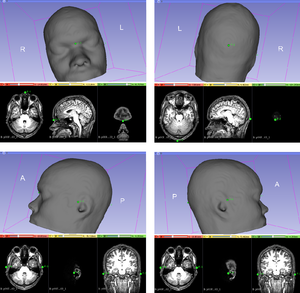
[[File:4 landmarks.png|thumb|right|Four anatomical landmarks are used for the essential positioning of the electrodes: the nasion, the inion, the pre auricular to the left ear and the pre auricular to the right ear. ]] | [[File:4 landmarks.png|thumb|right|Four anatomical landmarks are used for the essential positioning of the electrodes: the nasion, the inion, the pre auricular to the left ear and the pre auricular to the right ear. ]] | ||
| − | # 4 anatomical landmarks: (Sources Points) The list of fiducial points on the curve, since the "Create-and-place Fiducial" button (in green in the figure above). Four anatomical landmarks are used for the essential positioning of the electrodes (in this order!): | + | |
| − | #* 1/ The nasion | + | #4 anatomical landmarks: (Sources Points) The list of fiducial points on the curve, since the "Create-and-place Fiducial" button (in green in the figure above). Four anatomical landmarks are used for the essential positioning of the electrodes (in this order!): |
| − | #* 2/ The inion | + | #*1/ The nasion |
| − | #* 3/ The pre auricular to the left ear | + | #*2/ The inion |
| − | #* 4/ The pre auricular to the right ear | + | #*3/ The pre auricular to the left ear |
| − | # Input STL model: The model you use (after "use this mesh", the T1.stl created). | + | #*4/ The pre auricular to the right ear |
| − | # Press the button "Make 10-20 EEG system electrode" to draw the 10-20 EEG system via the Dijkstra's algorithm. | + | #Input STL model: The model you use (after "use this mesh", the T1.stl created). |
| − | #* The traditional T3P3 site according to the International 10–20 system of electroencephalogram was identified. | + | #Press the button "Make 10-20 EEG system electrode" to draw the 10-20 EEG system via the Dijkstra's algorithm. |
| + | #*The traditional T3P3 site according to the International 10–20 system of electroencephalogram was identified. | ||
*'''Project the stimulation site''' on the 10-20 system electrode distances and characterize it. | *'''Project the stimulation site''' on the 10-20 system electrode distances and characterize it. | ||
| − | # Stimulation Site placed: Place on the T1-weighted anatomical image the stimulation point that you want since the "Create-and-place Fiducial" button. Once this point given, click on 'Yes'. | + | |
| − | # Press the button "Project the stimulation site" to project the stimulation point on the scalp and find the 3 nearest electrodes around it. | + | #Stimulation Site placed: Place on the T1-weighted anatomical image the stimulation point that you want since the "Create-and-place Fiducial" button. Once this point given, click on 'Yes'. |
| − | #* Nearest electrode 1: The distance in centimeter between the first nearest electrode and the projected stimulation site. | + | #Press the button "Project the stimulation site" to project the stimulation point on the scalp and find the 3 nearest electrodes around it. |
| − | #* Nearest electrode 2: The distance in centimeter between the second nearest electrode and the projected stimulation site. | + | #*Nearest electrode 1: The distance in centimeter between the first nearest electrode and the projected stimulation site. |
| − | #* Nearest electrode 3: The distance in centimeter between the third nearest electrode and the projected stimulation site. | + | #*Nearest electrode 2: The distance in centimeter between the second nearest electrode and the projected stimulation site. |
| + | #*Nearest electrode 3: The distance in centimeter between the third nearest electrode and the projected stimulation site. | ||
| Line 100: | Line 109: | ||
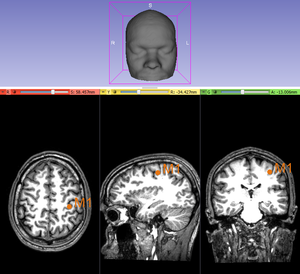
[[File:M1.png|thumb|Localization of the motor hand area via a knob on the precentral gyrus]] | [[File:M1.png|thumb|Localization of the motor hand area via a knob on the precentral gyrus]] | ||
| − | # M1 Point Placed: Place on the T1-weighted anatomical image a point targeting the human [https://en.wikipedia.org/wiki/Motor_cortex|motor motor cortex] since the "Create-and-place Fiducial" button. Once this point given, click on 'Yes'. Help via the [https://pdfs.semanticscholar.org/ba38/045e9f01ec4d128c5fbe5a46dc209fccaac4.pdf Yousry's method]. | + | #M1 Point Placed: Place on the T1-weighted anatomical image a point targeting the human [https://en.wikipedia.org/wiki/Motor_cortex|motor motor cortex] since the "Create-and-place Fiducial" button. Once this point given, click on 'Yes'. Help via the [https://pdfs.semanticscholar.org/ba38/045e9f01ec4d128c5fbe5a46dc209fccaac4.pdf Yousry's method]. |
| − | # Set the stimulation intensity of the resting motor threshold. | + | #Set the stimulation intensity of the resting motor threshold. |
| − | # Press the button "Correct the motor threshold" to correct the unadjusted motor threshold (rMT) in % stimulator output. | + | #Press the button "Correct the motor threshold" to correct the unadjusted motor threshold (rMT) in % stimulator output. |
| − | #* Two adjusted motor threshold (AdjMT%) in % stimulator output are given where SCDx is the scalp-to-cortex distance between the scalp and and the Stimulation Site, SCDm is the scalp-to-cortex distance between the scalp and M1. | + | #*Two adjusted motor threshold (AdjMT%) in % stimulator output are given where SCDx is the scalp-to-cortex distance between the scalp and and the Stimulation Site, SCDm is the scalp-to-cortex distance between the scalp and M1. |
| − | #* 1/ The first according to Stokes et al. Clin Neurophysiol 2007 <ref name="Stokes 2007">Stokes, M. G., Chambers, C. D., Gould, I. C., English, T., McNaught, E., McDonald, O., & Mattingley, J. B. (2007). Distance-adjusted motor threshold for transcranial magnetic stimulation. Clinical Neurophysiology, 118(7), 1617‑1625.</ref> , where [AdjMT% = 2,7*(SCDx - SCDm) + rMT] | + | #*1/ The first according to Stokes et al. Clin Neurophysiol 2007 <ref name="Stokes 2007">Stokes, M. G., Chambers, C. D., Gould, I. C., English, T., McNaught, E., McDonald, O., & Mattingley, J. B. (2007). Distance-adjusted motor threshold for transcranial magnetic stimulation. Clinical Neurophysiology, 118(7), 1617‑1625.</ref> , where [AdjMT% = 2,7*(SCDx - SCDm) + rMT] |
| − | #* 2/ The second according to Hoffman et al. Biol Psychiatry 2013 <ref name="Hoffman 2013">Hoffman, R. E., Wu, K., Pittman, B., Cahill, J. D., Hawkins, K. A., Fernandez, T., & Hannestad, J. (2013). Transcranial magnetic stimulation of Wernicke’s and Right homologous sites to curtail « voices »: a randomized trial. Biological Psychiatry, 73(10), 1008‑1014. </ref> , where [AdjMT% = 0.90*rMT*e0.036*(SCDx-SCDm)] | + | #*2/ The second according to Hoffman et al. Biol Psychiatry 2013 <ref name="Hoffman 2013">Hoffman, R. E., Wu, K., Pittman, B., Cahill, J. D., Hawkins, K. A., Fernandez, T., & Hannestad, J. (2013). Transcranial magnetic stimulation of Wernicke’s and Right homologous sites to curtail « voices »: a randomized trial. Biological Psychiatry, 73(10), 1008‑1014. </ref> , where [AdjMT% = 0.90*rMT*e0.036*(SCDx-SCDm)] |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|Information for Developers}} | {{documentation/{{documentation/version}}/extension-section|Information for Developers}} | ||
| − | The code is available at [https://github.com/ | + | The code is available at [https://github.com/FredericBr/SlicerGeodesic Github]. |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 117: | Line 126: | ||
{{documentation/{{documentation/version}}/module-footer}} | {{documentation/{{documentation/version}}/module-footer}} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| + | <references /> | ||
Latest revision as of 09:30, 16 October 2020
Home < Documentation < Nightly < Modules < GeodesicSlicer
|
For the latest Slicer documentation, visit the read-the-docs. |
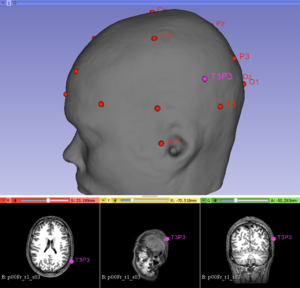
Introduction and Acknowledgements
|
The module has been developed based on ideas and feedbacks from the community. We would like to especially thank:
- Dr. Olivier Etard, M.D., Ph.D., CHU de Caen.
- Dr. Clément Nathou, M.D., Ph.D., CHU de Caen.
- Dr. Nicolas Delcroix, Ph.D., UMS 3408.
- Dr. Sonia Dollfus, M.D., Ph.D., CHU de Caen, header of ISTS.
- Dr. Csaba Pinter, MSc, Queen's University.
- Dr. Andras Lasso, Ph.D., Queen's University.
If you use this module, please cite the following article: [1] and [2].
| This extension is under the CeCill license, a copyleft license. |
Module Description
This module calculates geodesic path in 3D structure. Thanks to this geodesic path, this module can draw an EEG 10-20 system, determine the projected scalp stimulation site (MRI guided brain stimulation without the use of a neuronavigation System) and correct the rTMS resting motor threshold by correction factor.
Terminology
- Mesh A mesh or polygon mesh is a collection of vertices, edges and faces that defines the shape of a polyhedral object in 3D computer graphics and solid modeling.
- Shortest path In graph theory, the shortest path problem is the problem of finding a path between two vertices (or nodes) in a graph such that the sum of the weights of its constituent edges is minimized.
- 10-20 EEG system The International 10-20 system is commonly used for electroencephalogram (EEG) electrode placement and to correlate external skull locations with underlying cortical areas.[3]
Installation
- First, open 3D Slicer
- Open the Slicer Extensions from the icon on the menu bar
- Choose "Geodesic Slicer" module from the list of extensions and click "INSTALL" button.
- Once you restart 3D Slicer, the Geodesic Slicer module should show up on the Modules menu (under Informatics->Geodesic Slicer)
Use Cases
The overall goal is to allow users to find the shortest paths between nodes in a graph and via the Dijkstra's algorithm to make 10-20 system. This module can be used for:
- Stimulation in psychiatry: MRI guided brain stimulation without the use of a neuronavigation system.
- Surgery measurement.
- 3D printing.
Panels and their use
Create a mesh
A typical straightforward Geodesic Slicer workflow for consists of the following steps:
- Load a volume.nii (by Drag & Drop or the Add Data dialogue).
- Enter in the Geodesic Slicer module using either the toolbar or the Modules menu button.
- Press the button "Create a quick mesh" or "Create a mesh" (with filling holes smoothing, better for the next part but longer).
- Wait a moment.
- Go to Parameters to find the shortest path or Make 10-20 EEG system electrode section.
Parameters to find the shortest path
- Source points: The list of fiducial points on the curve, since the "Create-and-place Fiducial" button (in green in the figure above).
- Input STL model: The model you use (after "use this mesh", the T1.stl created).
- Find the shortest path: Calculate in centimeter the geodesic (shortest) path via the Dijkstra's algorithm.
- Draw the shortest path: Draw the Dijkstra's algorithm shortest path.
- Length (cm): The length of the current curve is shown in centimeter.
10-20 system electrode
- Run the Dijkstra's algorithm to make the 10-20 system electrode.
- 4 anatomical landmarks: (Sources Points) The list of fiducial points on the curve, since the "Create-and-place Fiducial" button (in green in the figure above). Four anatomical landmarks are used for the essential positioning of the electrodes (in this order!):
- 1/ The nasion
- 2/ The inion
- 3/ The pre auricular to the left ear
- 4/ The pre auricular to the right ear
- Input STL model: The model you use (after "use this mesh", the T1.stl created).
- Press the button "Make 10-20 EEG system electrode" to draw the 10-20 EEG system via the Dijkstra's algorithm.
- The traditional T3P3 site according to the International 10–20 system of electroencephalogram was identified.
- Project the stimulation site on the 10-20 system electrode distances and characterize it.
- Stimulation Site placed: Place on the T1-weighted anatomical image the stimulation point that you want since the "Create-and-place Fiducial" button. Once this point given, click on 'Yes'.
- Press the button "Project the stimulation site" to project the stimulation point on the scalp and find the 3 nearest electrodes around it.
- Nearest electrode 1: The distance in centimeter between the first nearest electrode and the projected stimulation site.
- Nearest electrode 2: The distance in centimeter between the second nearest electrode and the projected stimulation site.
- Nearest electrode 3: The distance in centimeter between the third nearest electrode and the projected stimulation site.
rTMS resting motor threshold- Correction factor
Calculate correction factors to adjust the rTMS dose for the treatment (according to the depth of the stimulation site).
- M1 Point Placed: Place on the T1-weighted anatomical image a point targeting the human motor cortex since the "Create-and-place Fiducial" button. Once this point given, click on 'Yes'. Help via the Yousry's method.
- Set the stimulation intensity of the resting motor threshold.
- Press the button "Correct the motor threshold" to correct the unadjusted motor threshold (rMT) in % stimulator output.
- Two adjusted motor threshold (AdjMT%) in % stimulator output are given where SCDx is the scalp-to-cortex distance between the scalp and and the Stimulation Site, SCDm is the scalp-to-cortex distance between the scalp and M1.
- 1/ The first according to Stokes et al. Clin Neurophysiol 2007 [4] , where [AdjMT% = 2,7*(SCDx - SCDm) + rMT]
- 2/ The second according to Hoffman et al. Biol Psychiatry 2013 [5] , where [AdjMT% = 0.90*rMT*e0.036*(SCDx-SCDm)]
Information for Developers
The code is available at Github.
References
- ↑ Briend F. et al., A new toolbox to compare NIBS localization method: Application for auditory hallucinations in schizophrenia. Schizophrenia Research, https://doi.org/10.1016/j.schres.2020.09.001
- ↑ Briend F. et al., GeodesicSlicer: a Slicer Toolbox for Targeting Brain Stimulation. Neuroinformatics. https://doi.org/10.1007/s12021-020-09457-9
- ↑ Jasper, H. (1958). The ten twenty electrode system of the international federation. Electroencephalography and Clinical Neurophysiology, 10, 371‑375.
- ↑ Stokes, M. G., Chambers, C. D., Gould, I. C., English, T., McNaught, E., McDonald, O., & Mattingley, J. B. (2007). Distance-adjusted motor threshold for transcranial magnetic stimulation. Clinical Neurophysiology, 118(7), 1617‑1625.
- ↑ Hoffman, R. E., Wu, K., Pittman, B., Cahill, J. D., Hawkins, K. A., Fernandez, T., & Hannestad, J. (2013). Transcranial magnetic stimulation of Wernicke’s and Right homologous sites to curtail « voices »: a randomized trial. Biological Psychiatry, 73(10), 1008‑1014.