Difference between revisions of "Documentation/Nightly/Extensions/LesionSpotlight"
Acsenrafilho (talk | contribs) |
Acsenrafilho (talk | contribs) m |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 27: | Line 27: | ||
[[Image:LesionSpotlight-logo.png|left]] | [[Image:LesionSpotlight-logo.png|left]] | ||
| − | This extension provides | + | This extension provides image segmentation and local image contrast enhancement approaches in order to detect abnormal white matter voxels signals in magnetic resonance images. At moment, there are available the '''LS Segmenter''' (specific for hyperintense Multiple Sclerosis lesion segmentation on T2-FLAIR images), '''LS Contrast Enhancement''' (specific to increase the contrast of abnormal voxels of T2-FLAIR images) and '''AFT Segmenter''' as a simple implementation of a recent automatic Multiple Sclerosis (MS) lesion segmentation<ref>DOI: 10.1016/j.cmpb.2014.04.006.</ref> approaches. The '''LS Segmenter''' module implements a T2-FLAIR hyperintense lesion segmentation based on the algorithm published in the paper<ref>DOI: 10.1007/s11517-017-1747-2</ref>. |
| − | '''NOTE''': The Logistic Contrast Enhancement, Weighted Enhancement Image Filter and Automatic FLAIR Threshold modules are only supporting CLI methods added in the extension to calculate specific parts of the segmentation procedure. For this reason, these modules are '''not''' supposed to be used alone. Please, use the '''LS Contrast Enhancer''' or '''LS Segmenter''' modules only. | + | '''NOTE''': The Logistic Contrast Enhancement, Weighted Enhancement Image Filter and Automatic FLAIR Threshold modules are '''only supporting CLI methods''' added in the extension in order to calculate specific parts of the segmentation procedure. For this reason, these modules are '''not''' supposed to be used alone. Please, use the '''LS Contrast Enhancer''' or '''LS Segmenter''' modules only. |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 35: | Line 35: | ||
*[[Documentation/{{documentation/version}}/Modules/LSSegmenter|LS Segmenter]] | *[[Documentation/{{documentation/version}}/Modules/LSSegmenter|LS Segmenter]] | ||
*[[Documentation/{{documentation/version}}/Modules/LSContrastEnhancer|LS Contrast Enhancer]] | *[[Documentation/{{documentation/version}}/Modules/LSContrastEnhancer|LS Contrast Enhancer]] | ||
| + | *[[Documentation/{{documentation/version}}/Modules/AFTSegmenter|AFT Segmenter]] | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Use Cases}} | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
Most frequently used for these scenarios: | Most frequently used for these scenarios: | ||
| − | * Use Case 1: Multiple Sclerosis (MS) | + | * Use Case 1: Hyperintense Multiple Sclerosis (MS) lesions segmentation |
| − | ** | + | **T2-FLAIR images are usually applied to MS diagnosis in order to detect hyperintense MS lesions. In this case, the '''LS Segmenter''' module can be useful. |
| − | * Use Case 2: Increase contrast in abnormal voxels in white matter | + | * Use Case 2: Increase contrast in abnormal voxels in white matter tissue |
| − | **There are some lesion segmentation approaches that | + | **There are some lesion segmentation approaches that rely on the voxel intensity level presented in the white matter tissue, where the '''LS Contrast Enhancer''' module can be helpful to increase the contrast between lesions and surrounding brain tissues (mainly normal-appearing white matter - NAWM). |
<gallery widths="300px" heights="300px" perrow="3"> | <gallery widths="300px" heights="300px" perrow="3"> | ||
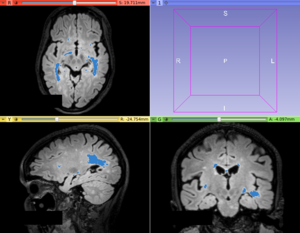
Image:T2FLAIR_patient.png|T2-FLAIR image from a MS patient | Image:T2FLAIR_patient.png|T2-FLAIR image from a MS patient | ||
| − | Image:T2FLAIR_patient_lesionLabel.png|T2-FLAIR hyperintense lesion segmentation provided by the LS Segmenter module | + | Image:T2FLAIR_patient_lesionLabel.png|T2-FLAIR hyperintense lesion segmentation provided by the '''LS Segmenter''' module |
| − | Image:Lesion3DRender.png| | + | Image:Lesion3DRender.png|MS lesions and White Matter 3D representation |
Image:T2FLAIR_beforeContrast.png|T2-FLAIR image with original lesion contrast | Image:T2FLAIR_beforeContrast.png|T2-FLAIR image with original lesion contrast | ||
| − | Image:T2FLAIR_afterContrast.png|T2-FLAIR image with hyperintense lesion contrast enhanced | + | Image:T2FLAIR_afterContrast.png|T2-FLAIR image with hyperintense MS lesion contrast enhanced by the '''LS Contrast Enhancer''' module |
</gallery> | </gallery> | ||
| Line 58: | Line 59: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|References}} | {{documentation/{{documentation/version}}/extension-section|References}} | ||
| − | * | + | * da Silva Senra Filho, A. C. (2017) ‘A hybrid approach based on logistic classification and iterative contrast enhancement algorithm for hyperintense multiple sclerosis lesion segmentation’, Medical & Biological Engineering & Computing. doi: 10.1007/s11517-017-1747-2. |
| + | * Cabezas, M., Oliver, A., Roura, E., Freixenet, J., Vilanova, J. C., Ramió-Torrentà, L., Rovira, À. and Lladó, X. (2014) ‘Automatic multiple sclerosis lesion detection in brain MRI by FLAIR thresholding’, Computer Methods and Programs in Biomedicine. Elsevier Ireland Ltd, 115(3), pp. 147–161. DOI: 10.1016/j.cmpb.2014.04.006. | ||
Latest revision as of 11:14, 6 December 2017
Home < Documentation < Nightly < Extensions < LesionSpotlight
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work was partially funded by CAPES and CNPq, a Brazillian Agencies. Information on CAPES can be obtained on the CAPES website and CNPq website. | |||||||||
|
Extension Description
This extension provides image segmentation and local image contrast enhancement approaches in order to detect abnormal white matter voxels signals in magnetic resonance images. At moment, there are available the LS Segmenter (specific for hyperintense Multiple Sclerosis lesion segmentation on T2-FLAIR images), LS Contrast Enhancement (specific to increase the contrast of abnormal voxels of T2-FLAIR images) and AFT Segmenter as a simple implementation of a recent automatic Multiple Sclerosis (MS) lesion segmentation[1] approaches. The LS Segmenter module implements a T2-FLAIR hyperintense lesion segmentation based on the algorithm published in the paper[2].
NOTE: The Logistic Contrast Enhancement, Weighted Enhancement Image Filter and Automatic FLAIR Threshold modules are only supporting CLI methods added in the extension in order to calculate specific parts of the segmentation procedure. For this reason, these modules are not supposed to be used alone. Please, use the LS Contrast Enhancer or LS Segmenter modules only.
Modules
Use Cases
Most frequently used for these scenarios:
- Use Case 1: Hyperintense Multiple Sclerosis (MS) lesions segmentation
- T2-FLAIR images are usually applied to MS diagnosis in order to detect hyperintense MS lesions. In this case, the LS Segmenter module can be useful.
- Use Case 2: Increase contrast in abnormal voxels in white matter tissue
- There are some lesion segmentation approaches that rely on the voxel intensity level presented in the white matter tissue, where the LS Contrast Enhancer module can be helpful to increase the contrast between lesions and surrounding brain tissues (mainly normal-appearing white matter - NAWM).
Similar Extensions
N/A
References
- da Silva Senra Filho, A. C. (2017) ‘A hybrid approach based on logistic classification and iterative contrast enhancement algorithm for hyperintense multiple sclerosis lesion segmentation’, Medical & Biological Engineering & Computing. doi: 10.1007/s11517-017-1747-2.
- Cabezas, M., Oliver, A., Roura, E., Freixenet, J., Vilanova, J. C., Ramió-Torrentà, L., Rovira, À. and Lladó, X. (2014) ‘Automatic multiple sclerosis lesion detection in brain MRI by FLAIR thresholding’, Computer Methods and Programs in Biomedicine. Elsevier Ireland Ltd, 115(3), pp. 147–161. DOI: 10.1016/j.cmpb.2014.04.006.
Information for Developers
| Section under construction. |
Repositories:
- Source code: GitHub repository
- Issue tracker: open issues and enhancement requests