Difference between revisions of "Documentation/Nightly/Extensions/SlicerDMRI"
(I think I've done my share of DMRI over the years... : )) |
|||
| (6 intermediate revisions by 2 users not shown) | |||
| Line 19: | Line 19: | ||
* NIH P41EB015902 (Neuroimaging Analysis Center) | * NIH P41EB015902 (Neuroimaging Analysis Center) | ||
* National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. | * National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. | ||
| − | Major Contributors: Lauren J. O'Donnell ({{collaborator|name|spl}}), Isaiah Norton (Brigham and Women's Hospital), Raul San Jose Estepar (Brigham and Women's Hospital), Carl-Fredrik Westin (Brigham and Women's Hospital)<br> | + | Major Contributors: Lauren J. O'Donnell ({{collaborator|name|spl}}), Isaiah Norton (Brigham and Women's Hospital), Raul San Jose Estepar (Brigham and Women's Hospital), Carl-Fredrik Westin (Brigham and Women's Hospital), Steve Pieper, Alex Yarmarkovich<br> |
License: [http://www.slicer.org/pages/LicenseText Slicer License] | License: [http://www.slicer.org/pages/LicenseText Slicer License] | ||
| + | |||
| + | Website: [http://slicerdmri.github.io SlicerDMRI] | ||
|} | |} | ||
| Line 46: | Line 48: | ||
{{documentation/{{documentation/version}}/extension-section|Modules}} | {{documentation/{{documentation/version}}/extension-section|Modules}} | ||
| − | *[[Documentation/ | + | * [[Documentation/Nightly/Modules/DiffusionWeightedVolumeMasking|Diffusion Brain Masking]]: Creates a brain mask from a diffusion weighted image volume |
| − | + | * [[Documentation/Nightly/Modules/DWIToDTIEstimation|Diffusion Tensor Estimation]]: Estimates the diffusion tensor model from diffusion weighted images. | |
| + | * [[Documentation/Nightly/Modules/DiffusionTensorScalarMeasurements|Diffusion Tensor Scalar Maps]]: Computes scalar measures from a diffusion tensor dataset. | ||
| + | * [[Documentation/Nightly/Modules/DWIConverter|DWIConverter]]: Converts diffusion weighted MR images in dicom series into Nrrd format for analysis in Slicer. | ||
| + | * [[Documentation/Nightly/Modules/ResampleDTIVolume|Resample DTIVolume]]: Implements DT image resampling through the use of itk Transforms. | ||
| + | * [[Documentation/Nightly/Modules/ResampleScalarVectorDWIVolume|Resample Scalar/Vector/DWI Volume]]: Implements image and vector-image resampling through the use of its Transforms. | ||
| + | * [[Documentation/Nightly/Modules/TractographyDICOMLoad|Tractography DICOM Load]]: Utility for DICOM TrackSet conversion to Slicer-style VTK tractography. | ||
| + | * [[Documentation/Nightly/Modules/TractographyDICOMSave|Tractography DICOM Save]]: Utility for conversion of Slicer-style VTK tractography to DICOM TrackSet. | ||
| + | * [[Documentation/Nightly/Modules/TractographyDisplay|Tractography Display]]: Allows the user to modify the display of diffusion tractography. | ||
| + | * [[Documentation/Nightly/Modules/FiberTractMeasurements|Tractography Measurements]]: Computes whole-fiber scalar measurements from a directory of VTK fiber bundle files | ||
| + | * [[Documentation/Nightly/Modules/FiberBundleLabelSelect|Tractography ROI Selection]]: Allows a user to select tracts passing or not passing through single or multiple labels. | ||
| + | * [[Documentation/Nightly/Modules/TractographyLabelMapSeeding|Tractography ROI Seeding]]: Seeds tractography from a Diffusion Tensor Image (DTI) within a region defined by a label map. | ||
| + | * [[Documentation/Nightly/Modules/TractographyInteractiveSeeding|Tractography Seeding]]: Allows interactive seeding of DTI fiber tracts starting from a list of fiducial points or vertices of a model, or a label map volume. | ||
| + | * [[Documentation/Nightly/Modules/FiberBundleToLabelMap|Tractography to Mask Image]]: Allows a user to set the specified label value in the label map at every vertex in each of the fibers in a bundle. | ||
| + | * [[Documentation/Nightly/Modules/UKFTractography|UKFTractography]]: Traces fibers in a DWI Volume using the multiple tensor unscented Kalman Filter methodology. | ||
| + | * DWI to Full Brain Tractography: | ||
{{documentation/{{documentation/version}}/extension-section|References}} | {{documentation/{{documentation/version}}/extension-section|References}} | ||
Latest revision as of 11:31, 12 October 2016
Home < Documentation < Nightly < Extensions < SlicerDMRI
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
| |||||||||
|
|}
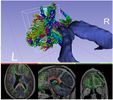
Extension Description
SlicerDMRI extension hosts various modules to facilitate
- processing and management of diffusion MRI image data
- utilizing diffusion MRI tractography for neuroscience studies and for planning image-guided interventions
- quantitative analysis of diffusion MRI data
Modules
- Diffusion Brain Masking: Creates a brain mask from a diffusion weighted image volume
- Diffusion Tensor Estimation: Estimates the diffusion tensor model from diffusion weighted images.
- Diffusion Tensor Scalar Maps: Computes scalar measures from a diffusion tensor dataset.
- DWIConverter: Converts diffusion weighted MR images in dicom series into Nrrd format for analysis in Slicer.
- Resample DTIVolume: Implements DT image resampling through the use of itk Transforms.
- Resample Scalar/Vector/DWI Volume: Implements image and vector-image resampling through the use of its Transforms.
- Tractography DICOM Load: Utility for DICOM TrackSet conversion to Slicer-style VTK tractography.
- Tractography DICOM Save: Utility for conversion of Slicer-style VTK tractography to DICOM TrackSet.
- Tractography Display: Allows the user to modify the display of diffusion tractography.
- Tractography Measurements: Computes whole-fiber scalar measurements from a directory of VTK fiber bundle files
- Tractography ROI Selection: Allows a user to select tracts passing or not passing through single or multiple labels.
- Tractography ROI Seeding: Seeds tractography from a Diffusion Tensor Image (DTI) within a region defined by a label map.
- Tractography Seeding: Allows interactive seeding of DTI fiber tracts starting from a list of fiducial points or vertices of a model, or a label map volume.
- Tractography to Mask Image: Allows a user to set the specified label value in the label map at every vertex in each of the fibers in a bundle.
- UKFTractography: Traces fibers in a DWI Volume using the multiple tensor unscented Kalman Filter methodology.
- DWI to Full Brain Tractography:
References
[1] O’Donnell LJ, Westin CF. An introduction to diffusion tensor image analysis. Neurosurgery clinics of North America. 2011 Apr 30;22(2):185-96.
Information for Developers
- SlicerSlicerDMRI organization page on github: https://github.com/SlicerDMRI