Difference between revisions of "Documentation/Nightly/Extensions/SlicerPathology"
(skeleton for coming documentation) |
|||
| (9 intermediate revisions by one other user not shown) | |||
| Line 20: | Line 20: | ||
{{documentation/{{documentation/version}}/module-introduction-end}} | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Module Description}} | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
| − | + | This extension provides tools for automatic and semi-automatic pathology image segmentation. | |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 32: | Line 31: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Tutorials}} | {{documentation/{{documentation/version}}/module-section|Tutorials}} | ||
| − | + | <embedvideo service="youtube">https://www.youtube.com/watch?v=n6RtJoU9nGQ</embedvideo> | |
| − | + | {| | |
| − | { | + | |[[File:SlicerPathologyScreenShot2.png|thumb|800px|Step 1 - Go to the SlicerPathology Extension.]] |
| − | { | + | |} |
| − | + | {| | |
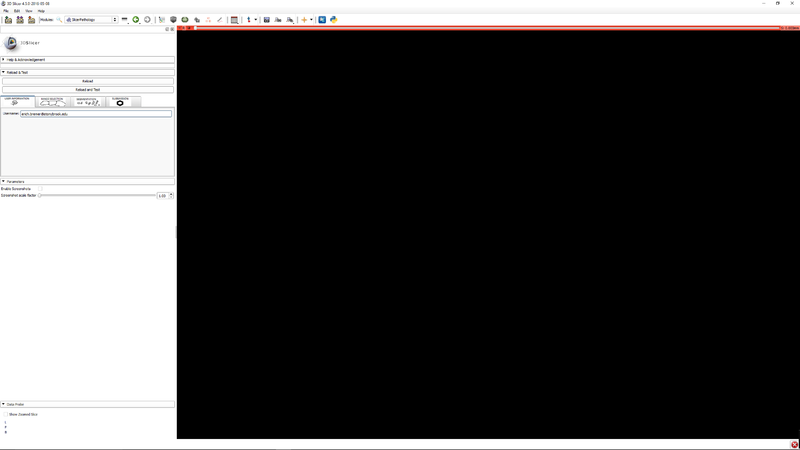
| + | |[[File:SlicerPathologyScreenShot3.png|thumb|800px|Step 2 - Click the user information tab. Enter your email address. This is used for identification of you in the file data files.]] | ||
| + | |} | ||
| + | {| | ||
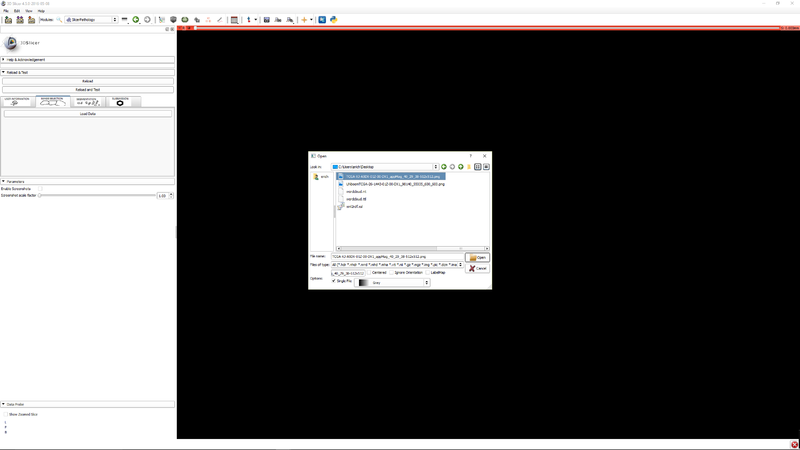
| + | |[[File:SlicerPathologyScreenShot4.png|thumb|800px|Step 3 - Click "Load data" to select an image.]] | ||
| + | |} | ||
| + | {| | ||
| + | |[[File:SlicerPathologyScreenShot5.png|thumb|800px|Step 4 - Click the button Quick TCGA Effect button to activate the effect.]] | ||
| + | |} | ||
| + | {| | ||
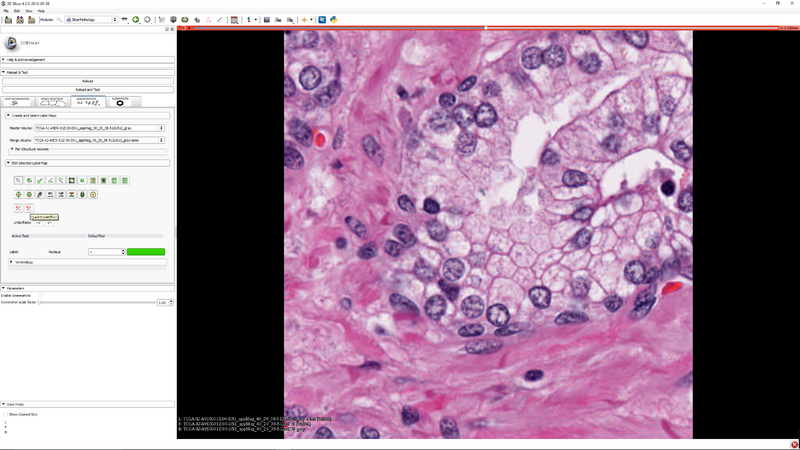
| + | |[[File:SlicerPathologyScreenShot6.png|thumb|800px|Step 5 - Click "Start TCGA Segmenter and adjust the five segmentation algorithm parameters as needed.]] | ||
| + | |} | ||
| + | {| | ||
| + | |[[File:SlicerPathologyScreenShot7.png|thumb|800px|Step 6 - you can select a subregion to speed up the parameter tuning process. Press "Y" to execute the segmenter on the subregion (if it was selected) or the whole image]] | ||
| + | |} | ||
| + | {| | ||
| + | |[[File:SlicerPathologyScreenShot8.png|thumb|800px|Step 7 - you can click "Clear Selection" to clear the selected sub-region so that the segmenter operates on the whole image.]] | ||
| + | |} | ||
| + | {| | ||
| + | |[[File:SlicerPathologyScreenShot9.png|thumb|800px|Step 8 - "Click the "Submission" tab so that you can save your label masks and related meta data. (meta data will be stored as JSON)]] | ||
| + | |} | ||
| + | {| | ||
| + | |[[File:SlicerPathologyScreenShot-json.png|thumb|800px|Step 9 - "This is a sample of the stored meta data. Notice the addition of your username for identification.]] | ||
| + | |} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|References}} | {{documentation/{{documentation/version}}/module-section|References}} | ||
* [http://qiicr.org Quantitative Image Informatics for Cancer Research (QIICR)] | * [http://qiicr.org Quantitative Image Informatics for Cancer Research (QIICR)] | ||
* [http://opencv.org Open Source Computer Vision] | * [http://opencv.org Open Source Computer Vision] | ||
| + | * [http://cancergenome.nih.gov/ The Cancer Genome Atlas] | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Information for Developers}} | {{documentation/{{documentation/version}}/module-section|Information for Developers}} | ||
| − | + | The source code for SlicerPathology is available at https://github.com/SBU-BMI/SlicerPathology | |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 20:54, 7 December 2016
Home < Documentation < Nightly < Extensions < SlicerPathology
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: SlicerPathology Contributor 1: Yi Gao |
Module Description
This extension provides tools for automatic and semi-automatic pathology image segmentation.
Use Cases
| Section under construction. |
Tutorials
References
- Quantitative Image Informatics for Cancer Research (QIICR)
- Open Source Computer Vision
- The Cancer Genome Atlas
Information for Developers
The source code for SlicerPathology is available at https://github.com/SBU-BMI/SlicerPathology