Difference between revisions of "Documentation/4.4/gif tutorial v3 2"
From Slicer Wiki
(Created page with '===Segmentation=== {| class="wikitable3" cellpadding="5px" |-align="center" |style="width:50%"|frame|600px |style="width:50%"|frame|600px |-valig…') |
|||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | |[[image:Neurosurgical Planning tutorial.png|700px|center]] | ||
| + | |||
| + | {| class="wikitable" style="border-spacing: 2px; border: 1px solid darkgray;" | ||
| + | ! scope="col" | Table of Contents | ||
| + | |- | ||
| + | |[[Documentation/4.4/gif tutorial v3 1|1. Loading DTI and Baseline Data]] | ||
| + | |- | ||
| + | |[[Documentation/4.4/gif tutorial v3 3|3. Tractography exploration of peritumoral white matter fibers]] | ||
| + | |- | ||
| + | |[[Documentation/4.4/gif tutorial v3 4|4. Tractography exploration of the ipsilateral and contralateral side]] | ||
| + | |} | ||
| + | |||
===Segmentation=== | ===Segmentation=== | ||
| Line 37: | Line 49: | ||
|11. Position the mouse over the pin icon, and select the eye icon to view the all the slices in the 3D viewer. | |11. Position the mouse over the pin icon, and select the eye icon to view the all the slices in the 3D viewer. | ||
|} | |} | ||
| + | |||
| + | |||
| + | |||
| + | <div style="text-align: center;">'''[[Documentation/4.4/gif tutorial v3 3|Next Step]]'''</div> | ||
| + | <div style="text-align: center;">'''[[Documentation/4.4/gif tutorial v3 1|Previous Step]]'''</div> | ||
Latest revision as of 20:27, 20 July 2015
Home < Documentation < 4.4 < gif tutorial v3 2|
| Table of Contents |
|---|
| 1. Loading DTI and Baseline Data |
| 3. Tractography exploration of peritumoral white matter fibers |
| 4. Tractography exploration of the ipsilateral and contralateral side |
Segmentation
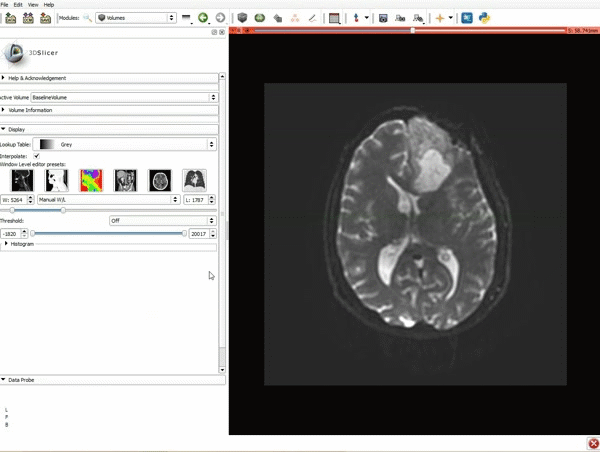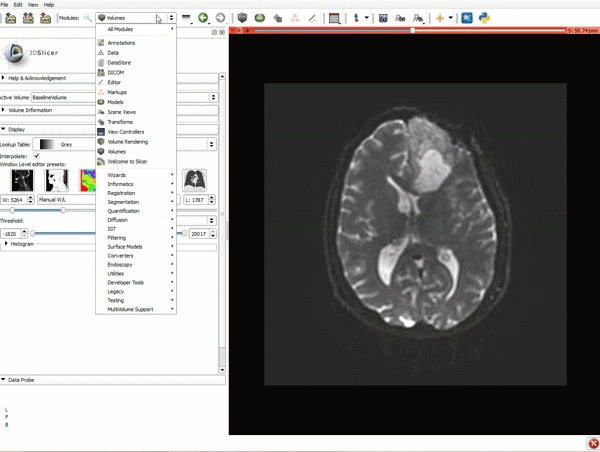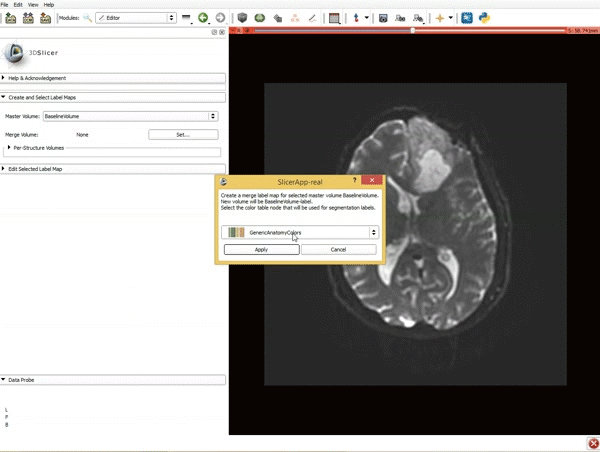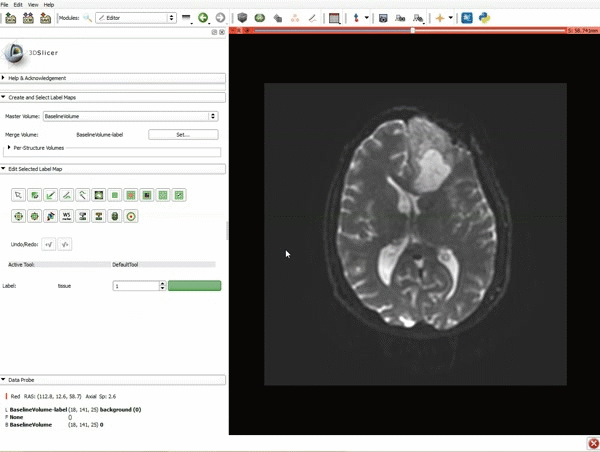
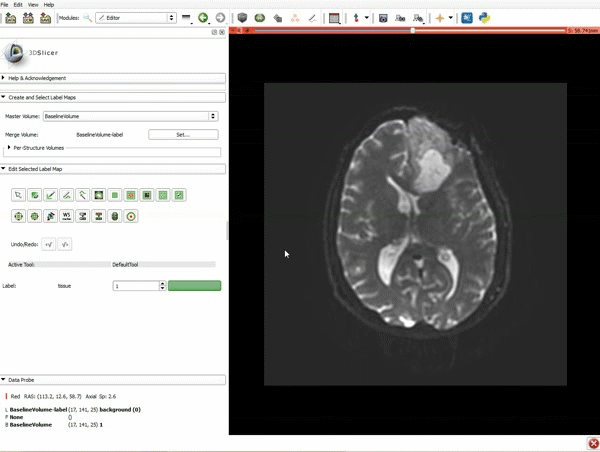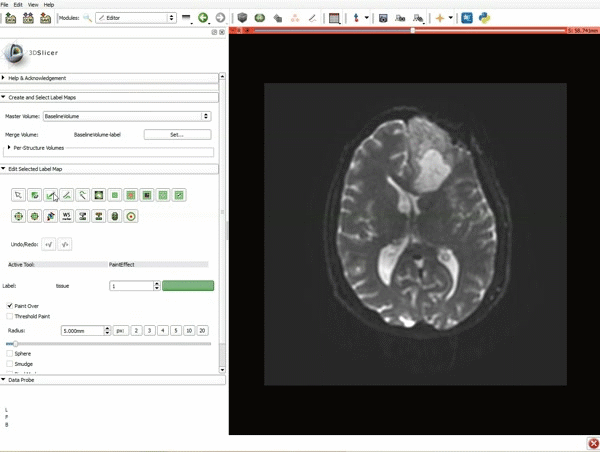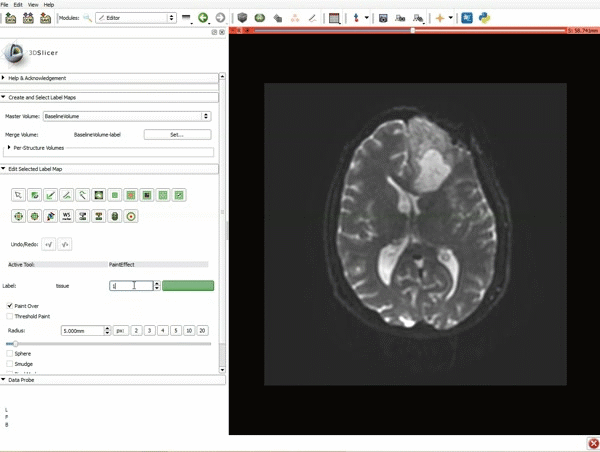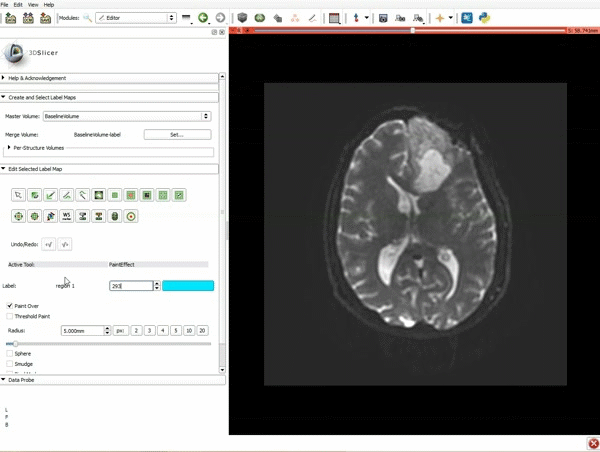
| 1. a) Click on the Modules menu and select the module Editor. b)The ‘SlicerApp-real’ window appears. Select the label map GenericAnatomyColors and click Apply. | 2. a) Select the PaintEffect b) Scroll down the Editor module and choose color #293 for the region 1 label |
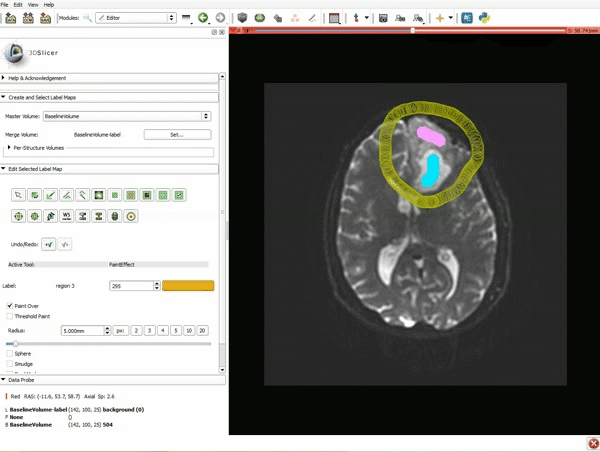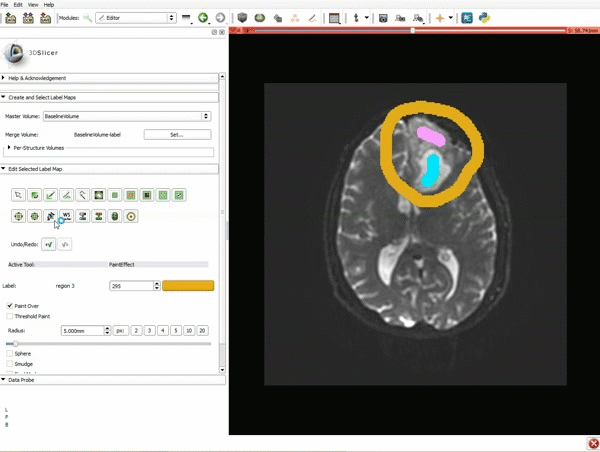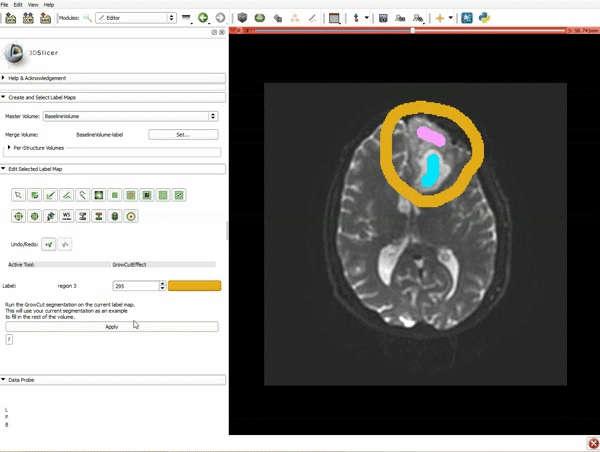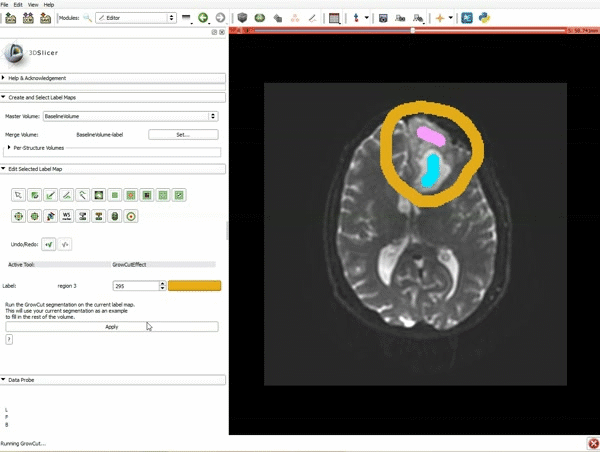
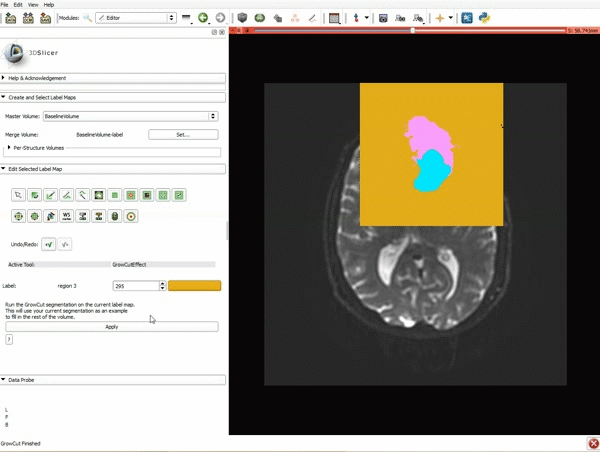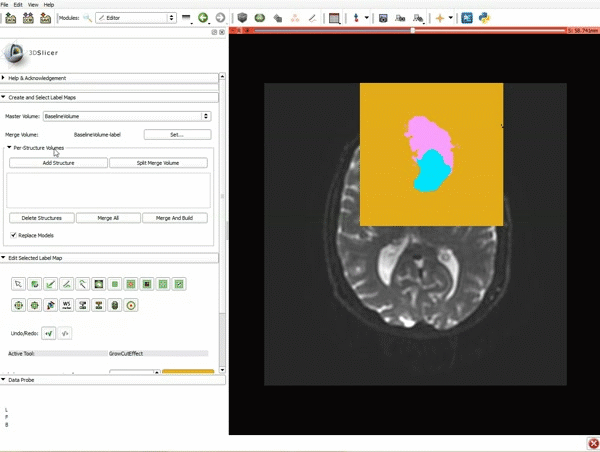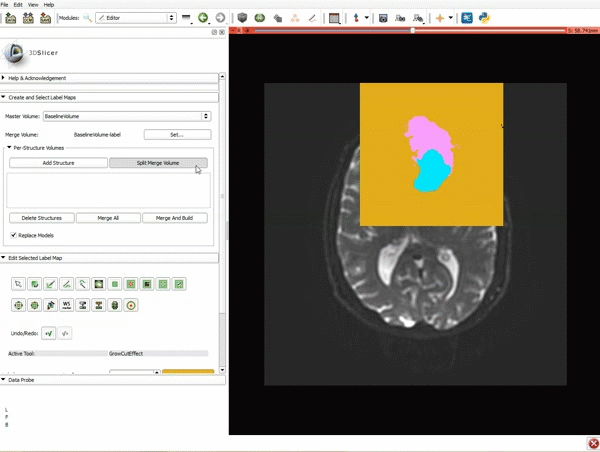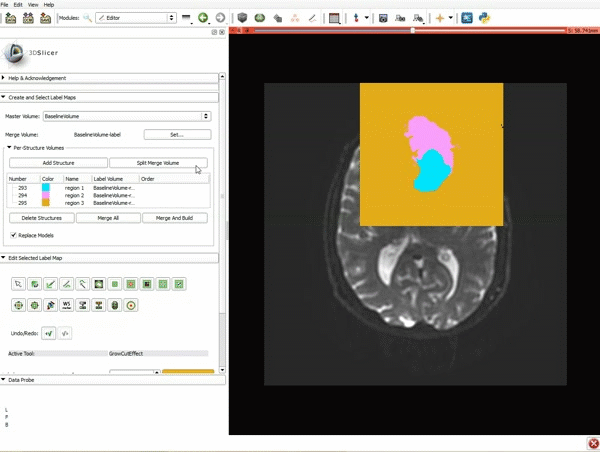
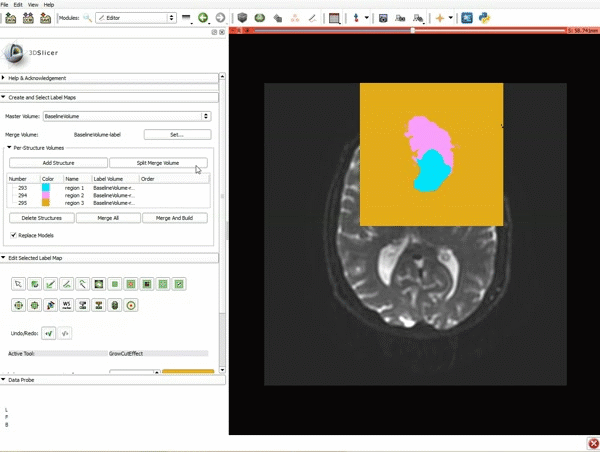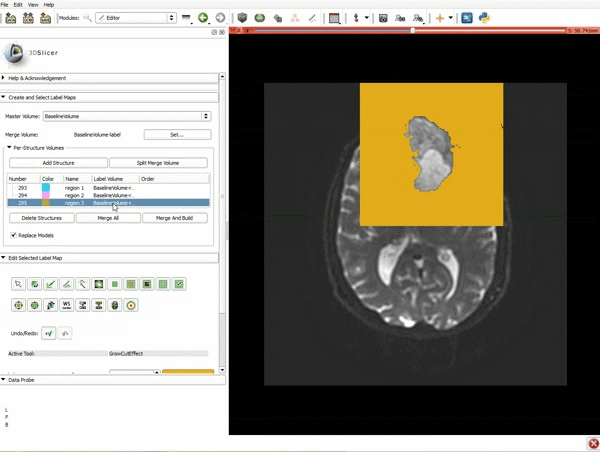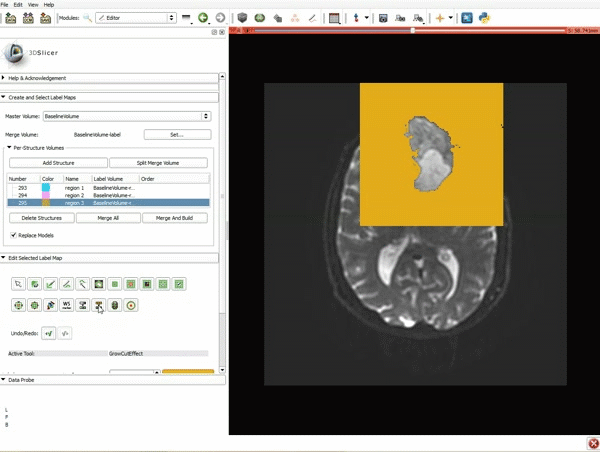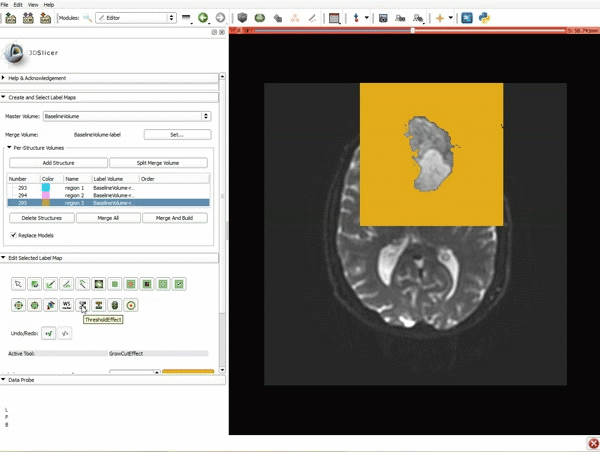
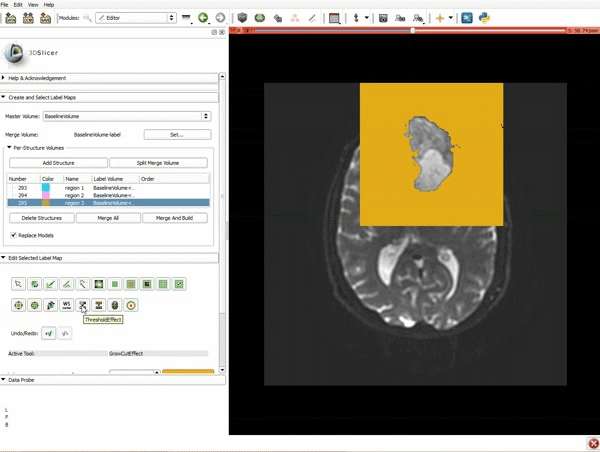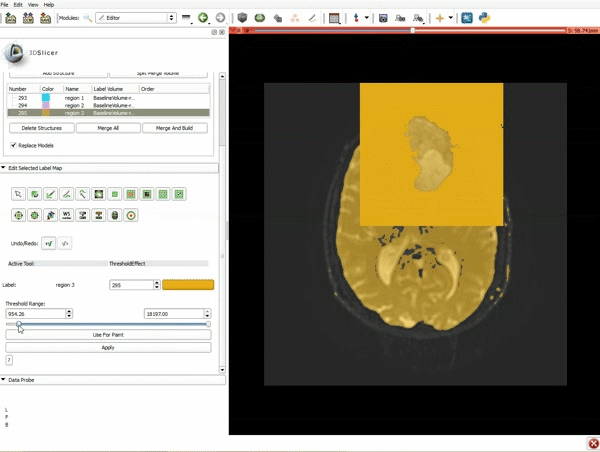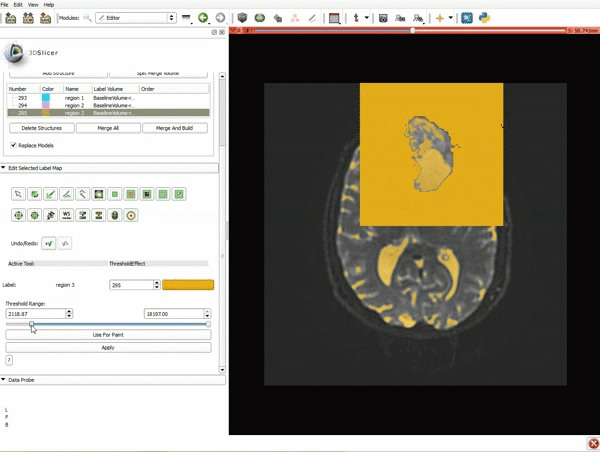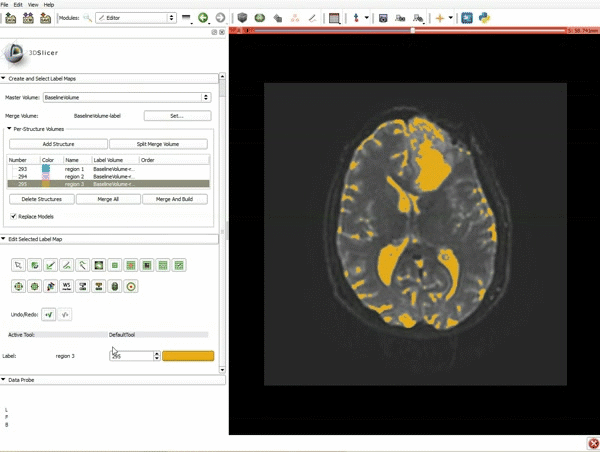
| 3. a) In a single motion, draw a short line in the cystic part of the tumor b) Select color #294 for the mass label and, again in a single motion, draw a short line in the solid part of the tumor c) Select color #295 for the region 3 and draw a circle around the tumor | 4. Select the GrowCutEffect tool and then click Apply |
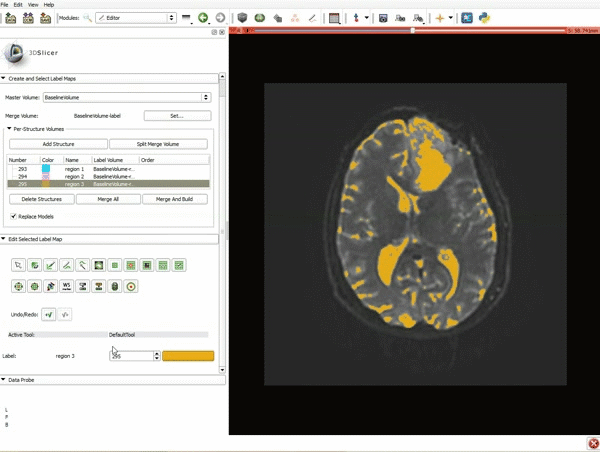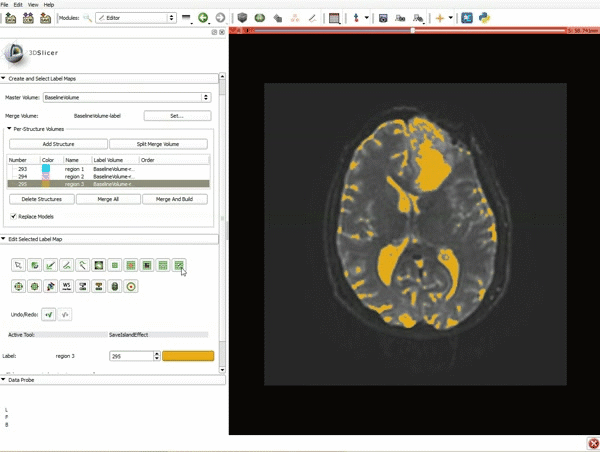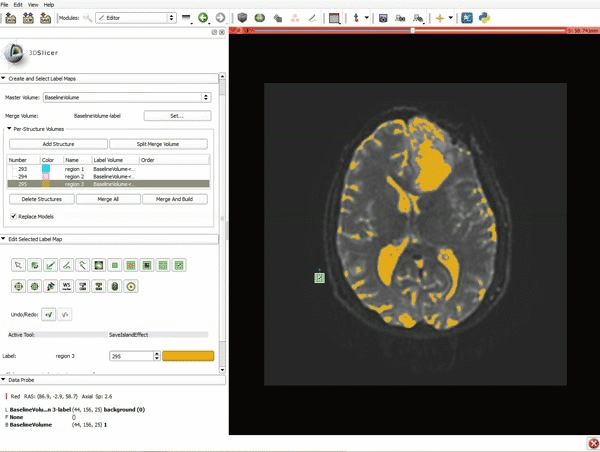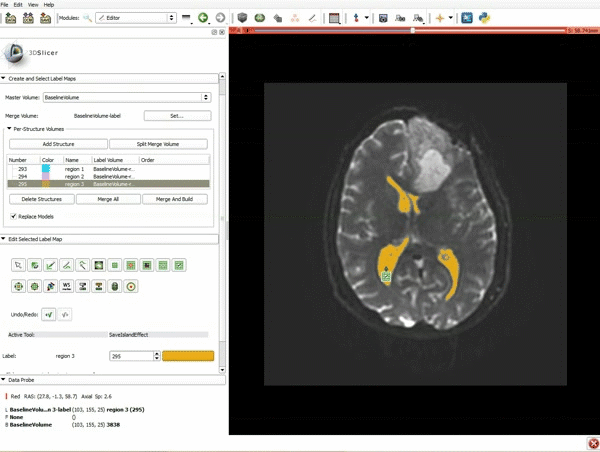
| 5. Scroll up the Editor menu and select the tab Per-Structure Volumes. Then click Split Merge Volume. | 6. a) Select the volume BaselineVolumeregion_3-label so that it is highlighted and that the yellow region is visible in the viewer b) Select the ThresholdEffect tool |
| 7. Scroll down the Editor module and set the lower Threshold Range to 1700 and click Apply. | 8. a) Select the SaveIslandEffect in the Editor module b) With the SaveIslandEffect tool equipped, click in the occipital horn of the ventricle. |
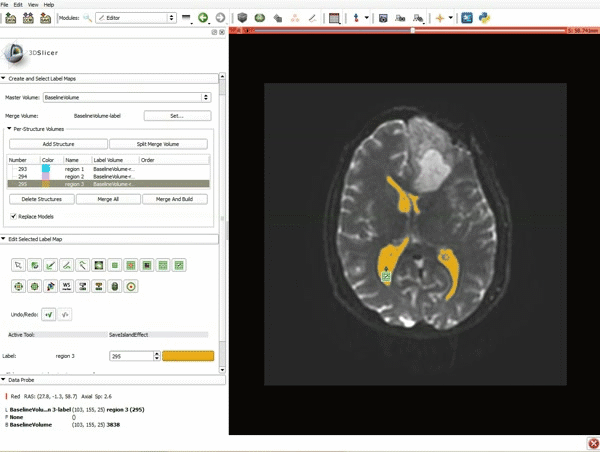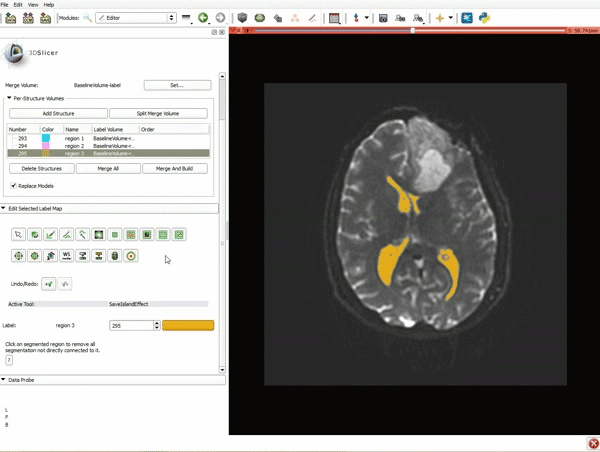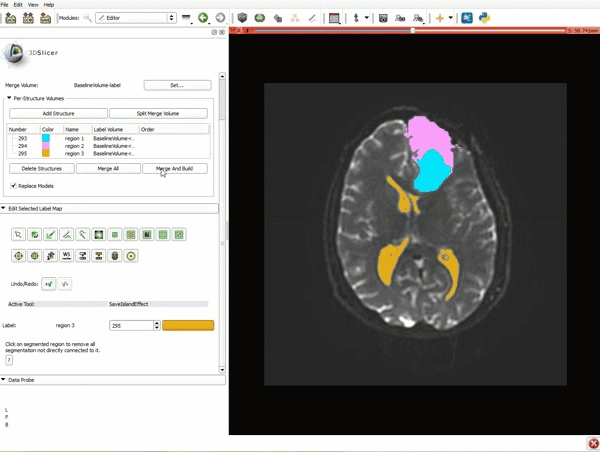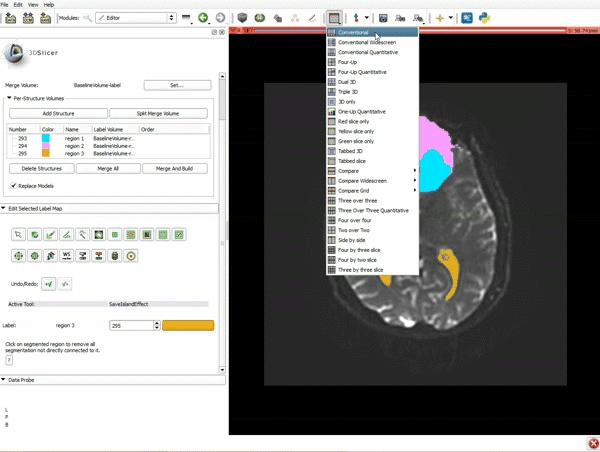
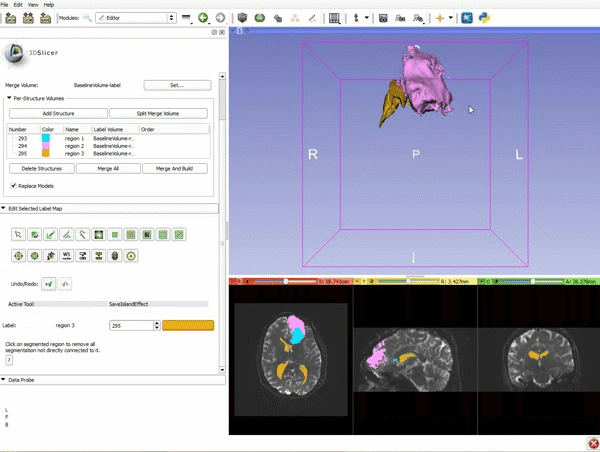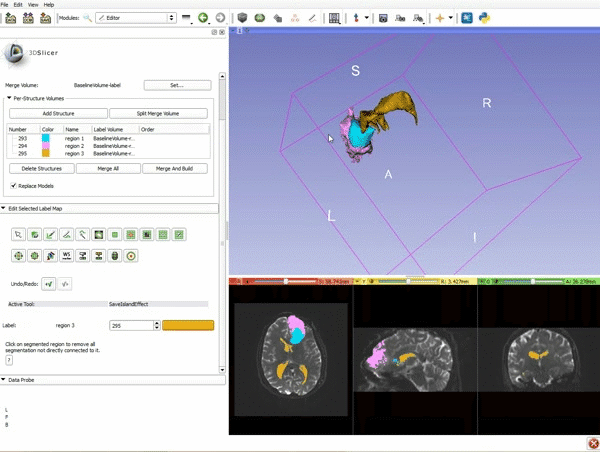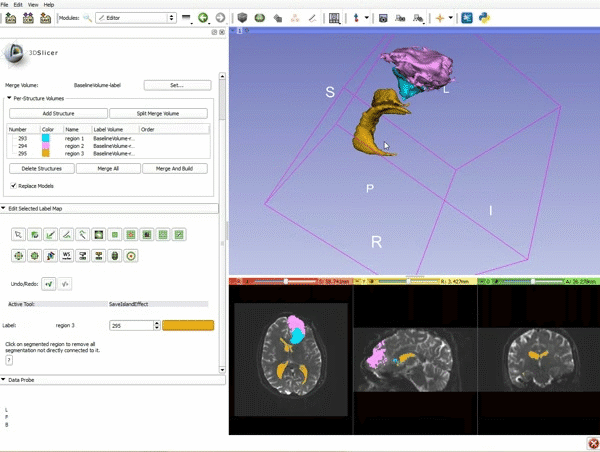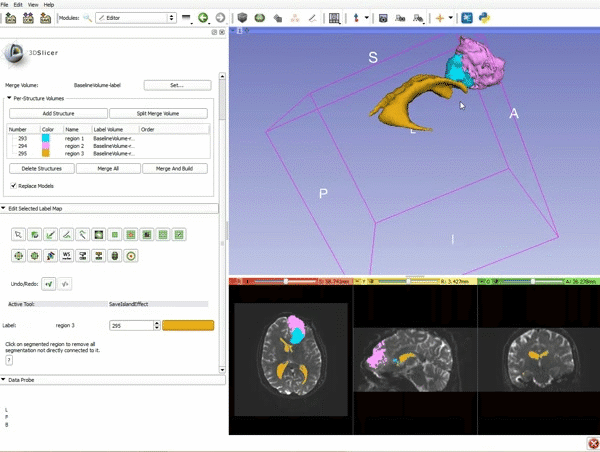
| 9. a) Scroll back up and click on Merge and Build to merge the different label maps, and generate the 3D models of the tumor and ventricles using a Marching Cubes algorithm. b) Click on the Layout menu and select Conventional. | 10. Slicer displays the 3D surface reconstructions of the ventricles, and solid and cystic parts of the tumor. |
| 11. Position the mouse over the pin icon, and select the eye icon to view the all the slices in the 3D viewer. |