Difference between revisions of "Slicer3:Module:QueryAtlas"
| (2 intermediate revisions by 2 users not shown) | |||
| Line 7: | Line 7: | ||
|} | |} | ||
| + | <flash>file=QAQdec.swf|width=1000|height=800|quality=best</flash> | ||
| + | <flash>file=QAFIPSFreeSurfer.swf|width=1000|height=800|quality=best</flash> | ||
== General Information == | == General Information == | ||
| Line 134: | Line 136: | ||
Source code is available here: | Source code is available here: | ||
| − | http:// | + | http://viewvc.slicer.org/viewcvs.cgi/trunk/Modules/QueryAtlas/ |
Links to documentation generated by doxygen: | Links to documentation generated by doxygen: | ||
Latest revision as of 14:33, 9 February 2009
Home < Slicer3:Module:QueryAtlasQueryAtlas
<flash>file=QAQdec.swf|width=1000|height=800|quality=best</flash>
<flash>file=QAFIPSFreeSurfer.swf|width=1000|height=800|quality=best</flash>
General Information
Module Type & Category
Type: Interactive
Category: Data visualization and inspection
Authors & Collaborators
Steve Pieper, Greg Brown, David Kennedy, Maryanne Martone, Jyl Boline, Burak Ozyurt,Wendy Plesniak, Michael Halle, Anna Tang, and Florin Talos
Related presentations
QueryAtlas Slides w/o Movies (ppt) for Oct07 BIRN AHM
QueryAtlas Slides including Movies (zip) for Oct07 BIRN AHM
QueryAtlas FIPS / FreeSurfer (combined morphology and functional analyses) data query movie (zip)
QueryAtlas Qdec data (population statistics on inflated brain) query movie (zip)
Module Description
The Query Atlas is a 3D Slicer module that allows the use of atlas-based anatomical representations, linked to underlying semantic descriptions, as an interactive 3D spatial reference for viewing experimental results and for performing information searches to study and interpret them. Query Atlas also provides a simple search gateway to web-based life sciences databases, and streamlines the information search process by generating appropriate queries for selected (currently mostly publication-related) search targets from a single query interface. Its underlying mapping of atlas-based anatomical structure through ontological definitions allows information searches based on different nomenclatures to be conducted.
The Query Atlas supports a number of different use cases:
- performing a hypothesis-driven information search in the context of particular experimental results
- searching for information to explain an unexpected post-hoc finding,
- discovering semantically related terms for anatomy of interest (possibly independent of atlas or data visualization), and
- learning about anatomy, anatomical hierarchies, different nomenclatures, and supporting information.
This project aims to address several challenges associated with scientific inquiry or study in the context of anatomical morphology:
- inefficiencies in searching distributed information resources,
- general “information overload”, particularly during early-stage research,
- lack of explicit links between semantically-related information resources,
and to provide an integrated environment for exploring experiment results, anatomical atlas and related multi-modality and multi-scale scientific information. The QueryAtlas currently supports the FreeSurfer atlas-based brain segmentation and its local naming conventions, and can be extended to support other volumetric or surface-based datasets labeled with atlas-based annotations.
The current version of the QueryAtlas module can be used to:
- Load volumetric analyses from XCEDE compliant or XNAT databases;
- Visualize volumes, models and parametric map overlays.
- Annotate models and labelmaps with atlas-based anatomy labels
- Map those local anatomy names to symantically compatible terms used in the larger biomedical informatics (neuroinformatics) community
- Formulate queries in the context of particular study results (fMRI activations or group statistics, for example)
- View, manage and save collections of query results
Usage
Example data
Download FIPS/FreeSurfer (XCEDE catalog) example data: QueryAtlasExampleData.zip .
Download Qdec (XNAT) example data: TEST5.qdec .
Description of Features and Use
- Pop-up help:
All widgets (and icons) in the module should display balloon help when the mouse hovers over them. The pop-up message will explain the widget's functionality.
- Scene Setup panel:
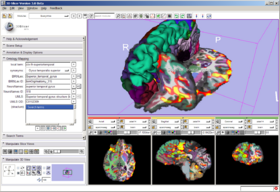
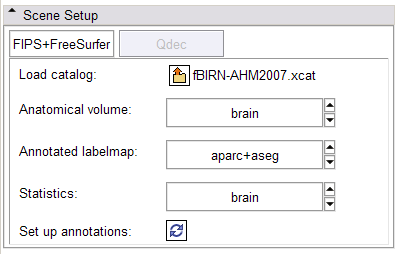
In the FIPS/FreeSurfer tab, load FIPS/FreeSurfer morphological and functional analyses from Xcede 2.0 catalog files, arrange data in the foreground, background and label layer of the Slice Viewers through the interface, and set up annotations on the datasets. Complete datasets should include a high resolution anatomical scan (brain.mgz), a label map generated by FreeSurfer's automatic parcellation (aparc+aseg.mgz), the pial surface models (lh.pial and rh.pial) and accompanying annotations (lh.aparc.annot and rh.aparc.annot), an example functional dataset used in the FIPS analysis (example_func.nii) and resulting brain activation statistics of interest (zstat.nii, zfstat.nii), and the matrix FreeSurfer uses to register the example functional and anatomical image (anat2efx.register.dat). For best behavior, before loading a FIPS/FreeSurfer dataset, delete other annotated datasets in the scene, or close the scene entirely.
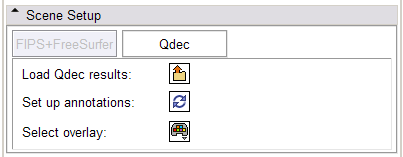
In the Qdec tab, load a Qdec population statistics analysis results file (file.qdec) extracted from an XNAT archive, select from among the scalar overlays representing the statistics in the study, and set up annotations on the dataset. As mentioned above, for best behavior, before loading a Qdec dataset, delete other annotated datasets in the scene, or close the scene entirely.
- Annotation & Display Options panel:
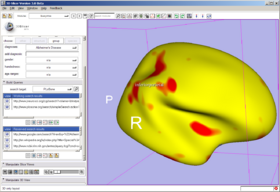
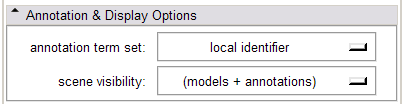
In the Annotation & Display Options, select among termsets to be used to generate the interactive annotations in the 3D Viewer (local terms, BIRNLex Strings, NeuroNames Strings, IBVD terms, and UMLS Concept Names). Anatomical regions that are mapped from the local termset to any of the others will be annotated on mouse-over; if no translation of the local term is available, no annotation will be displayed. The Scene Visibility menu can be used to turn on and off annotations in the 3D viewer, and to toggle model visibility, which can be useful for viewing slice planes and label map or statistical overlays that a model may occlude.
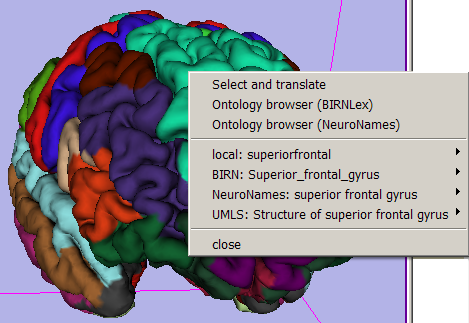
- 3D Viewer's Context Menu:
This menu, available by right-clicking in the 3D viewer when mousing over an annotated anatomical region, offers 'quick search' functionality for each translated term, and populates the Ontology Mapping panel described below. Note: quick (and all) web search actions require that you use Mozilla Firefox, and point Slicer to its executable. You can do that in Slicer's Application Settings Interface (which can be launched from the View->Application Settings menu).
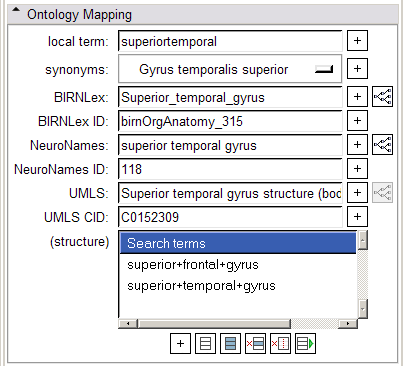
- Ontology Mapping panel:
The widgets in the Ontology Mapping panel are populated by the context menu available in Slicer's Main Viewer on right-click. The top menu choice, "Select and Translate" will select the current annotated region and map it through all termsets in the controlled vocabulary. The "synonyms" menubutton can be used to display a menu of synonyms for the annotated region, as specified by the NeuroNames ontology. Clicking the "+" button by any of the terms will add that term to the structure terms list box below. These terms can be selected, deleted, or added to the structure term list in the "Search Terms" panel described below.
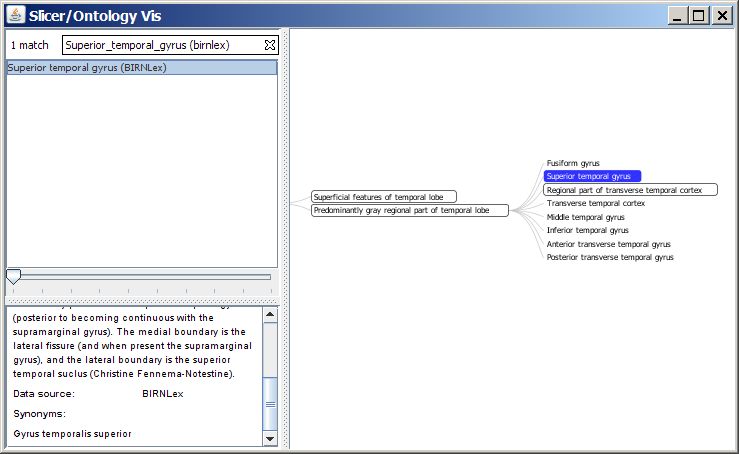
- SPL Ontology Browser:
Selecting the hierarchy buttons for BIRNLex or NeuroNames will launch the SPL Ontology Browser and display the anatomical hierarchy for that term as an interactive information visualization, and will provide some additional description for that term as well. This browser is written in Java and is based on the open-source Prefuse toolkit for infoviz. It contains a subset of the BIRNLex ontology and all of NeuroNames, including synonyms. You will need to have Java 1.5+ installed on your machine in order to launch and run the browser.
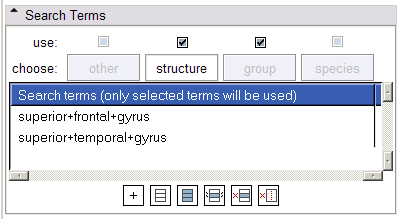
- Search Terms panel:
The tabs in this panel allow the specification of categories of search terms (Other, Structure, Population and Species). Structure terms inherit those terms sent from the Ontology Mapping panel, and new terms may also be specified. The use of the four categories of terms in formulating links can be toggled via the checkbox above each category's tab. Additionally, only terms in the Structure and Other tabs which are selected will be used in formulating links.
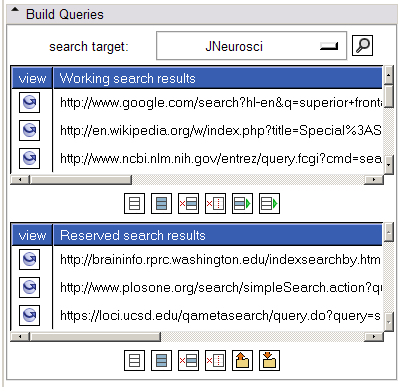
- Build Queries panel:
This panel allows a search target (or all search targets) to be selected. Selecting the 'search' button will form appropriate links for the selected target(s) and display unique ones in the "working search results" listbox. Each link may be inspected by clicking on the "view" icon to its left; this action raises the web browser and displays the url in a new tab. Useful links may be reserved to the "reserved search results" listbox and others may be deleted.
It's especially useful to collapse all other GUI panels except for the Search Terms and Build Queries panel -- this makes it easy to modify terms and generate a new search.
At the end of a session, reserved links may be saved to a Firefox bookmark file for future reference. Firefox bookmark files from previous searches may also be loaded back in to the interface to continue building an information collection from a past study.
Development
Software Dependencies
- Mozilla Firefox 2: Can be downloaded from http://www.mozilla.com/en-US/
- Java 1.5+: Can be downloaded from http://java.com/en/download/index.jsp
Known bugs
Follow this link to the Slicer3 bug tracker to see known bugs, or to report a new one:
http://na-mic.org/Mantis/main_page.php
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing:
http://na-mic.org/Mantis/main_page.php
Source code & documentation
Source code is available here:
http://viewvc.slicer.org/viewcvs.cgi/trunk/Modules/QueryAtlas/
Links to documentation generated by doxygen:
http://www.na-mic.org/Slicer/Documentation/Slicer3/html/
More Information
Controlled Vocabulary
This excel spreadsheet contains the mapping of FreeSurfer segmentation and parcellation names to BIRNLex, NeuroNames, UMLS, and IBVD. This is a work in progress; any corrections to the document are welcome.
Acknowledgement
This research was supported by Grant 5 MOI RR 000827 to the FIRST BIRN and Grant 1 U24 RR021992 to the FBIRN Biomedical Informatics Research Network (BIRN, http://www.nbirn.net), that is funded by the National Center for Research Resources (NCRR) at the National Institutes of Health (NIH). This work was also supported by NA-MIC, NAC, NCIGT. NeuroNames ontology and URI resources are provided courtesy of BrainInfo, University of Washington.